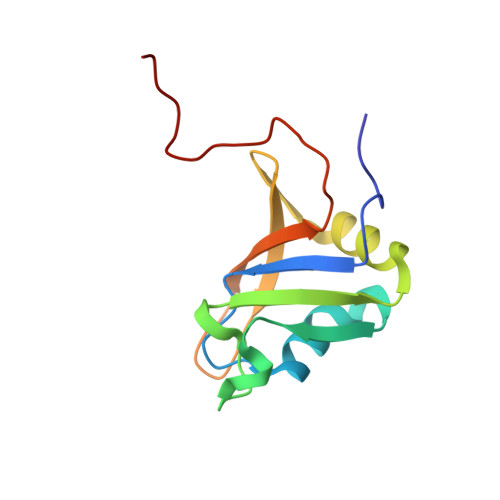
Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Tsuda, K., Kuwasako, K., Nagata, T., Takahashi, M., Kigawa, T., Kobayashi, N., Guntert, P., Shirouzu, M., Yokoyama, S., Muto, Y.(2014) Proteins 82: 2879-2886
- PubMed: 25066254
- DOI: https://doi.org/10.1002/prot.24651
- Primary Citation of Related Structures:
2RUG - PubMed Abstract:
The family of cytoplasmic polyadenylation element binding proteins CPEB1, CPEB2, CPEB3, and CPEB4 binds to the 3'-untranslated region (3'-UTR) of mRNA, and plays significant roles in mRNA metabolism and translation regulation. They have a common domain organization, involving two consecutive RNA recognition motif (RRM) domains followed by a zinc finger domain in the C-terminal region. We solved the solution structure of the first RRM domain (RRM1) of human CPEB3, which revealed that CPEB3 RRM1 exhibits structural features distinct from those of the canonical RRM domain. Our structural data provide important information about the RNA binding ability of CPEB3 RRM1.
- RIKEN Systems and Structural Biology Center, Tsurumi-ku, Yokohama, 230-0045, Japan; Division of Structural and Synthetic Biology, RIKEN Center for Life Science Technologies, Tsurumi-ku, Yokohama, 230-0045, Japan.
Organizational Affiliation: