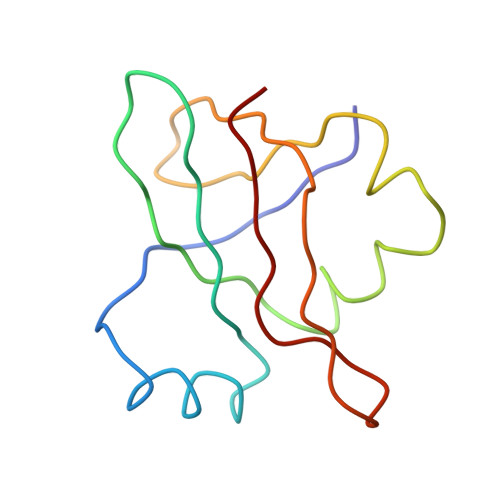
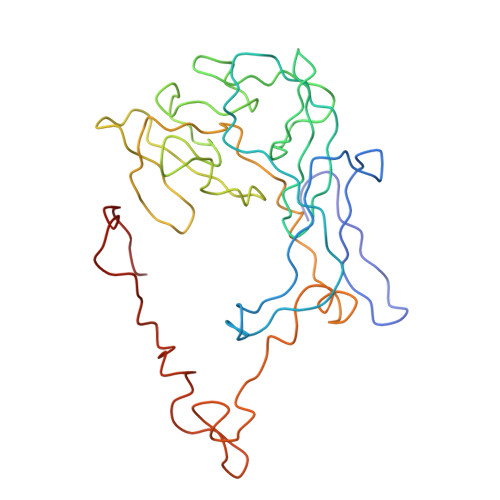
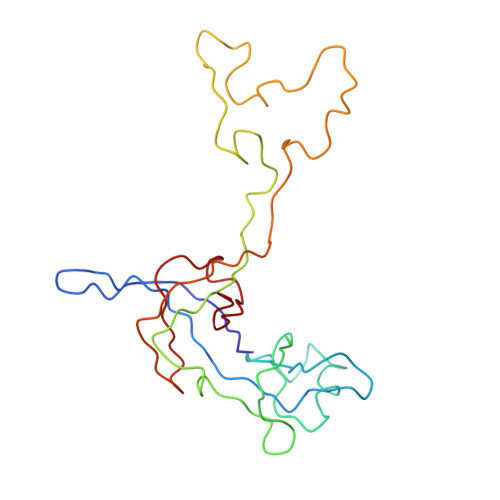
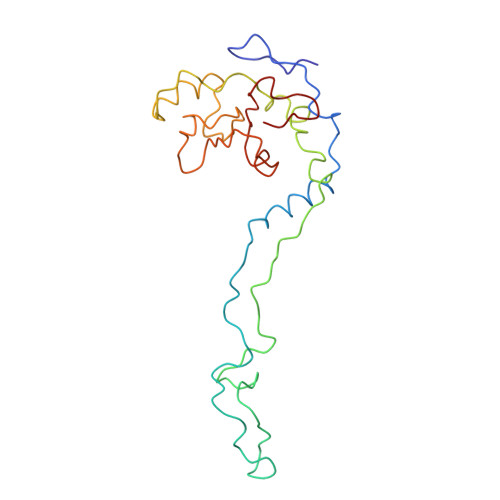
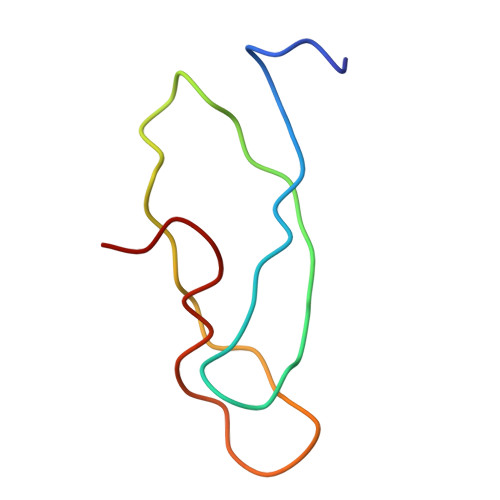
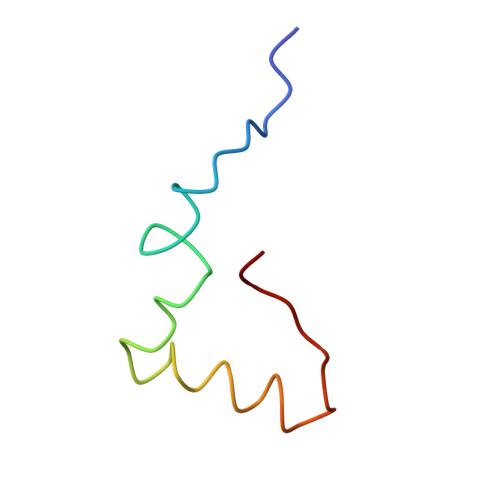
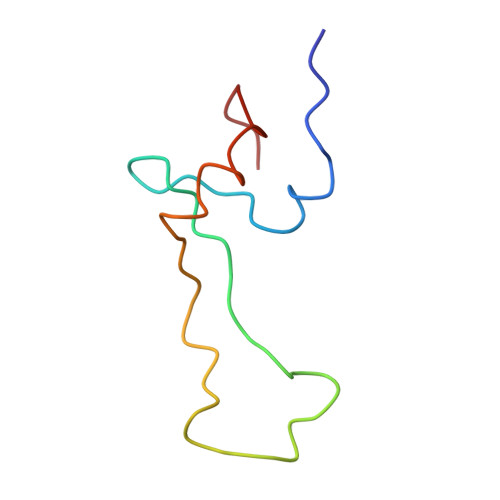
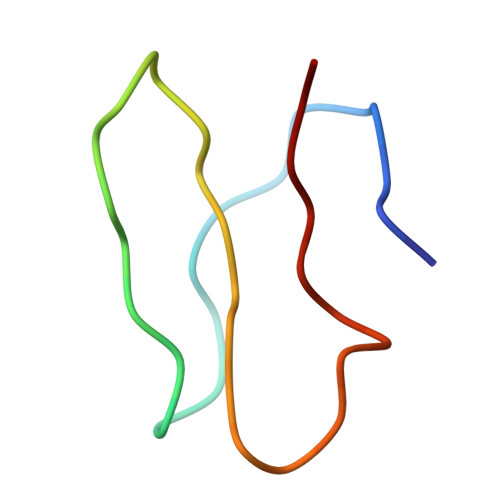
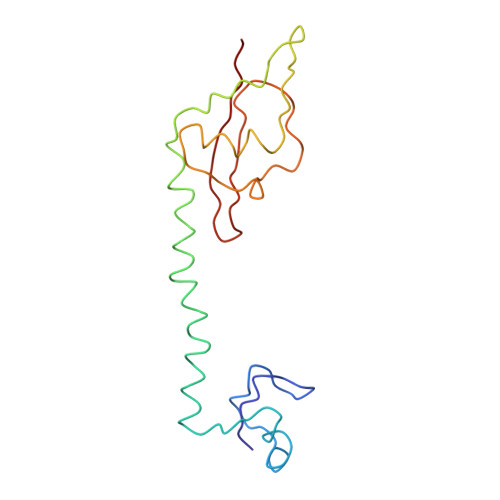
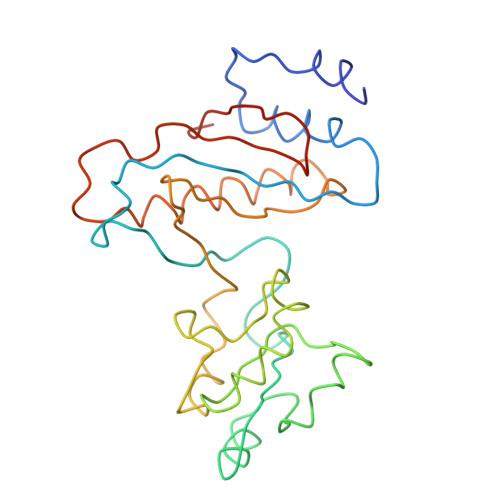
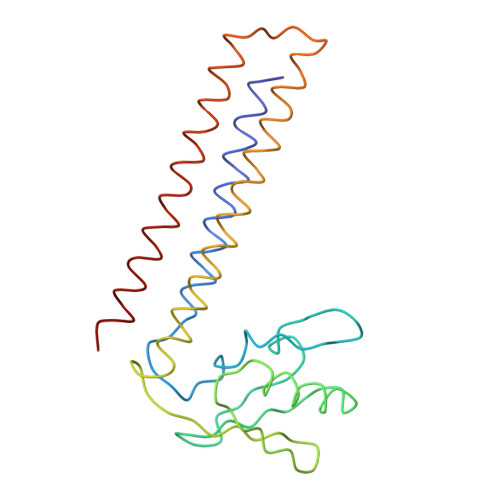
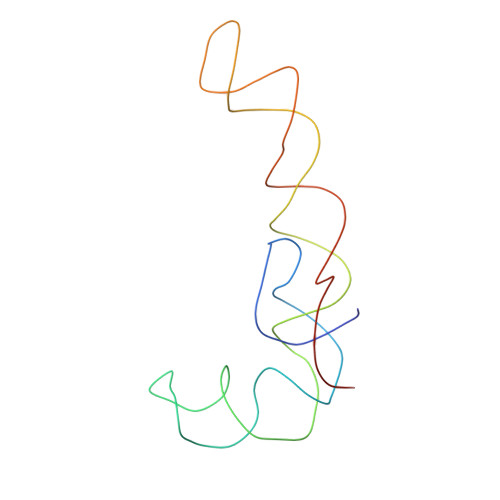Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
Gao, N., Zavialov, A.V., Ehrenberg, M., Frank, J.(2007) J Mol Biology 374: 1345-1358
- PubMed: 17996252
- DOI: https://doi.org/10.1016/j.jmb.2007.10.021
- Primary Citation of Related Structures:
2RDO - PubMed Abstract:
After termination of protein synthesis, the bacterial ribosome is split into its 30S and 50S subunits by the action of ribosome recycling factor (RRF) and elongation factor G (EF-G) in a guanosine 5'-triphosphate (GTP)-hydrolysis-dependent manner. Based on a previous cryo-electron microscopy study of ribosomal complexes, we have proposed that the binding of EF-G to an RRF-containing posttermination ribosome triggers an interdomain rotation of RRF, which destabilizes two strong intersubunit bridges (B2a and B3) and, ultimately, separates the two subunits. Here, we present a 9-A (Fourier shell correlation cutoff of 0.5) cryo-electron microscopy map of a 50S x EF-G x guanosine 5'-[(betagamma)-imido]triphosphate x RRF complex and a quasi-atomic model derived from it, showing the interaction between EF-G and RRF on the 50S subunit in the presence of the noncleavable GTP analogue guanosine 5'-[(betagamma)-imido]triphosphate. The detailed information in this model and a comparative analysis of EF-G structures in various nucleotide- and ribosome-bound states show how rotation of the RRF head domain may be triggered by various domains of EF-G. For validation of our structural model, all known mutations in EF-G and RRF that relate to ribosome recycling have been taken into account. More importantly, our results indicate a substantial conformational change in the Switch I region of EF-G, suggesting that a conformational signal transduction mechanism, similar to that employed in transfer RNA translocation on the ribosome by EF-G, translates a large-scale movement of EF-G's domain IV, induced by GTP hydrolysis, into the domain rotation of RRF that eventually splits the ribosome into subunits.
- Howard Hughes Medical Institute, Wadsworth Center, Empire State Plaza, Albany, NY 12201-0509, USA.
Organizational Affiliation: