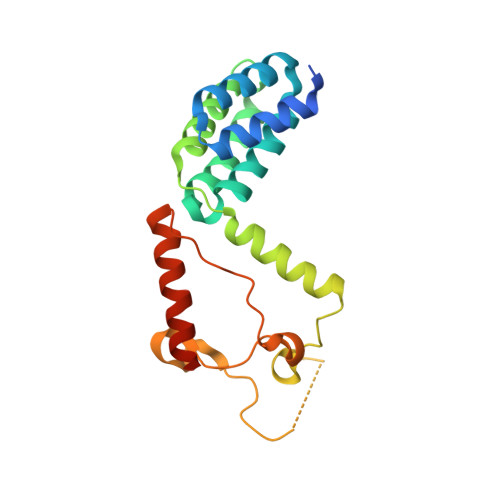
Crystal Structure of the C-Terminal Domain of AAA ATPase from Enterococcus faecium.
Ramagopal, U.A., Patskovsky, Y., Bonanno, J.B., Shi, W., Toro, R., Meyer, A.J., Rutter, M., Wu, B., Groshong, C., Gheyi, T., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.