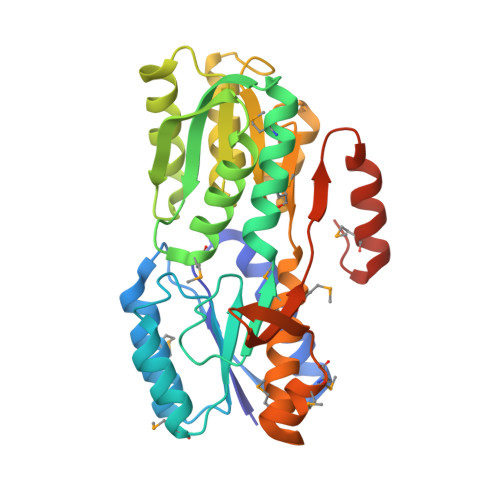
Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Palani, K., Kumaran, D., Burley, S.K., Swaminathan, S.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1460-1464
- PubMed: 23192024
- DOI: https://doi.org/10.1107/S1744309112045241
- Primary Citation of Related Structures:
2QVC - PubMed Abstract:
ABC transport systems have been characterized in organisms ranging from bacteria to humans. In most bacterial systems, the periplasmic component is the primary determinant of specificity of the transport complex as a whole. Here, the X-ray crystal structure of a periplasmic glucose-binding protein (GBP) from Thermotoga maritima determined at 2.4 Å resolution is reported. The molecule consists of two similar α/β domains connected by a three-stranded hinge region. In the current structure, a ligand (β-D-glucose) is buried between the two domains, which have adopted a closed conformation. Details of the substrate-binding sites revealed features that determine substrate specificity. In toto, ten residues from both domains form eight hydrogen bonds to the bound sugar and four aromatic residues (two from each domain) stabilize the substrate through stacking interactions.
- Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA.
Organizational Affiliation: