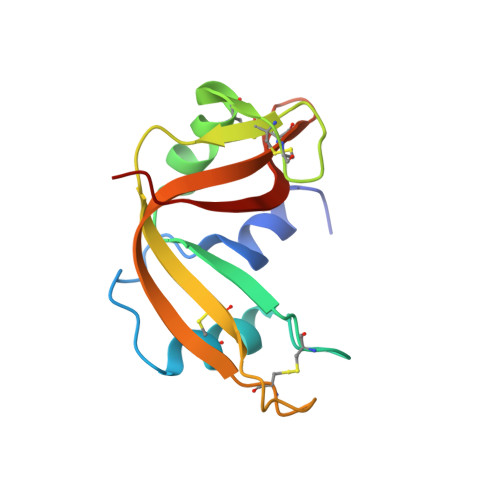
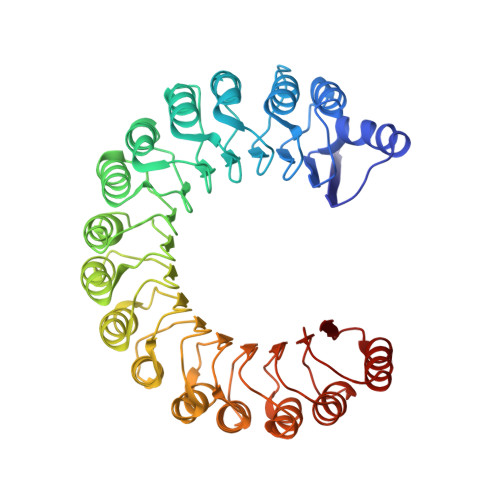
Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein.
Johnson, R.J., McCoy, J.G., Bingman, C.A., Phillips Jr., G.N., Raines, R.T.(2007) J Mol Biology 368: 434-449
- PubMed: 17350650
- DOI: https://doi.org/10.1016/j.jmb.2007.02.005
- Primary Citation of Related Structures:
1Z7X, 2Q4G - PubMed Abstract:
The ribonuclease inhibitor protein (RI) binds to members of the bovine pancreatic ribonuclease (RNase A) superfamily with an affinity in the femtomolar range. Here, we report on structural and energetic aspects of the interaction between human RI (hRI) and human pancreatic ribonuclease (RNase 1). The structure of the crystalline hRI x RNase 1 complex was determined at a resolution of 1.95 A, revealing the formation of 19 intermolecular hydrogen bonds involving 13 residues of RNase 1. In contrast, only nine such hydrogen bonds are apparent in the structure of the complex between porcine RI and RNase A. hRI, which is anionic, also appears to use its horseshoe-shaped structure to engender long-range Coulombic interactions with RNase 1, which is cationic. In accordance with the structural data, the hRI.RNase 1 complex was found to be extremely stable (t(1/2)=81 days; K(d)=2.9 x 10(-16) M). Site-directed mutagenesis experiments enabled the identification of two cationic residues in RNase 1, Arg39 and Arg91, that are especially important for both the formation and stability of the complex, and are thus termed "electrostatic targeting residues". Disturbing the electrostatic attraction between hRI and RNase 1 yielded a variant of RNase 1 that maintained ribonucleolytic activity and conformational stability but had a 2.8 x 10(3)-fold lower association rate for complex formation and 5.9 x 10(9)-fold lower affinity for hRI. This variant of RNase 1, which exhibits the largest decrease in RI affinity of any engineered ribonuclease, is also toxic to human erythroleukemia cells. Together, these results provide new insight into an unusual and important protein-protein interaction, and could expedite the development of human ribonucleases as chemotherapeutic agents.
- Department of Biochemistry, University of Wisconsin-Madison, Madison, WI 53706-1544, USA.
Organizational Affiliation: