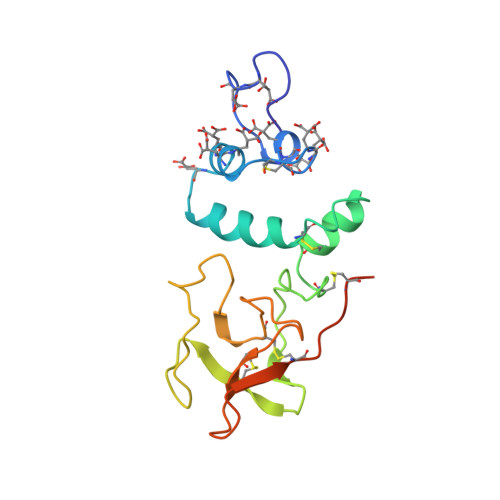
The Ca2+ ion and membrane binding structure of the Gla domain of Ca-prothrombin fragment 1.
Soriano-Garcia, M., Padmanabhan, K., de Vos, A.M., Tulinsky, A.(1992) Biochemistry 31: 2554-2566
- PubMed: 1547238
- DOI: https://doi.org/10.1021/bi00124a016
- Primary Citation of Related Structures:
2PF2 - PubMed Abstract:
The structure of Ca-prothrombin fragment 1 (residues 1-156 prothrombin) has been solved and refined at 2.2-A resolution by X-ray crystallographic methods. The first two-thirds of the Gla domain (residues 1-48) and two carbohydrate chains (approximately 5 kDa) are disordered in crystals of apo-fragment 1. When crystals are grown in the presence of Ca2+ ions, the Gla domain exhibits a well-defined structure binding seven Ca2+ ions, but the carbohydrate is still disordered. Even so, the crystallographic R factor reduced to 0.171. The folding of the Gla domain is dominated by 9-10 turns of three different alpha-helices. These turns produce two internal carboxylate surfaces composed of Gla side chains. A polymeric array of five Ca2+ ions separated by about 4.0 A intercalates between the carboxylate surfaces. The coordination of the Ca2+ ions with Gla carboxylate oxygen atoms and water molecules leads to distorted polyhedral arrangements with mu-oxo bridges in a highly complex array that most likely orchestrates the folding of the domain. The overall mode of interaction of the Ca2+ ions is new and different from any Ca2+ ion-protein interactions heretofore observed or described. The fluorescence quenching event observed upon Ca2+ ion binding is due to a disulfide-pi-electron interaction that causes a 100 degrees reorientation of Trp42 of the Gla domain. The Ca2+ ion interaction also affords the N-terminus protection from acetylation because the latter is buried in the folded structure and makes hydrogen-bonding salt bridges with Gla17, Gla21, and Gla27. The Gla domain and its trailing disulfide unit associate intimately and together give rise to a domain-like structure. Electrostatic potential calculations indicate that the Gla domain is very electronegative. Since most of the carboxylate oxygen atoms of Gla residues are involved in Ca2+ ion binding, leaving only a few for bridging Ca2+ ion-phospholipid interactions, the role of bridging Ca2+ ions might be generally unspecific, with Ca2+ ions simply intervening between the negative Gla domain and negative head groups of the membrane surface. The folding of the kringle structure in apo- and Ca-fragment 1 is essentially the same. However, the Ser36-Ala47 helix of the Gla domain pivots around Cys48, shifting by approximately 30 degrees, and the helix encroaches on the kringle producing some concomitant changes. These might be related to the protection of carbohydrate carrying Asn101 from acetylation in the Ca-fragment 1 structure.
- Department of Chemistry, Michigan State University, East Lansing 48824.
Organizational Affiliation: