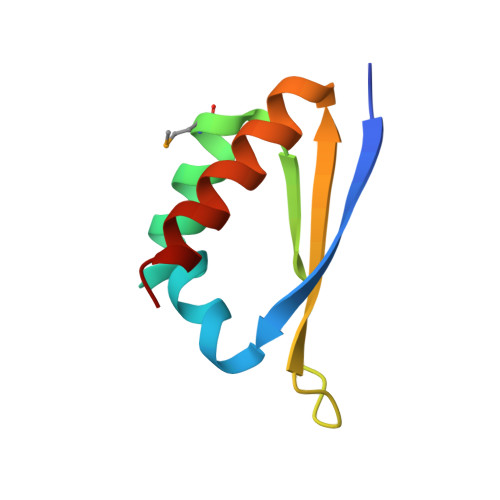
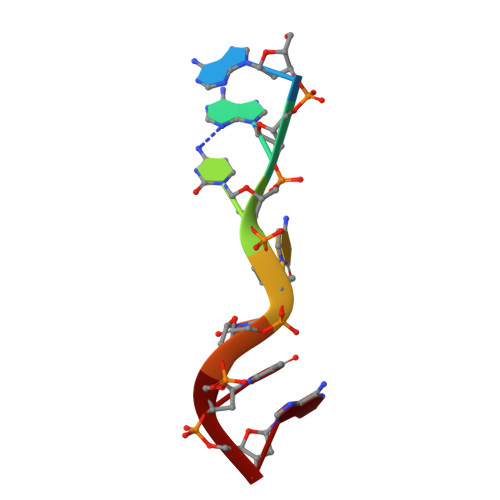
Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution.
Fenn, S., Du, Z., Lee, J.K., Tjhen, R., Stroud, R.M., James, T.L.(2007) Nucleic Acids Res 35: 2651-2660
- PubMed: 17426136
- DOI: https://doi.org/10.1093/nar/gkm139
- Primary Citation of Related Structures:
2P2R - PubMed Abstract:
KH (hnRNP K homology) domains, consisting of approximately 70 amino acid residues, are present in a variety of nucleic-acid-binding proteins. Among these are poly(C)-binding proteins (PCBPs), which are important regulators of mRNA stability and posttranscriptional regulation in general. All PCBPs contain three different KH domains and recognize poly(C)-sequences with high affinity and specificity. To reveal the molecular basis of poly(C)-sequence recognition, we have determined the crystal structure, at 1.6 A resolution, of PCBP2 KH3 domain in complex with a 7-nt DNA sequence (5'-AACCCTA-3') corresponding to one repeat of the C-rich strand of human telomeric DNA. The domain assumes a type-I KH fold in a betaalphaalphabetabetaalpha configuration. The protein-DNA interface could be studied in unprecedented detail and is made up of a series of direct and water-mediated hydrogen bonds between the protein and the DNA, revealing an especially dense network involving several structural water molecules for the last 2 nt in the core recognition sequence. Unlike published KH domain structures, the protein crystallizes without protein-protein contacts, yielding new insights into the dimerization properties of different KH domains. A nucleotide platform, an interesting feature found in some RNA molecules, was identified, evidently for the first time in DNA.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, California 94143-2280, USA.
Organizational Affiliation: