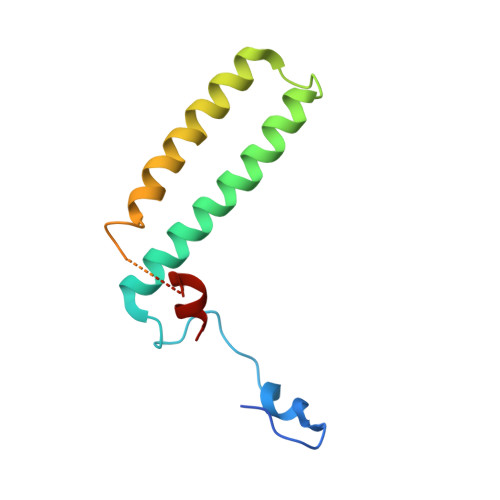
Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer.
Kostelansky, M.S., Schluter, C., Tam, Y.Y., Lee, S., Ghirlando, R., Beach, B., Conibear, E., Hurley, J.H.(2007) Cell 129: 485-498
- PubMed: 17442384
- DOI: https://doi.org/10.1016/j.cell.2007.03.016
- Primary Citation of Related Structures:
2P22 - PubMed Abstract:
The endosomal sorting complex required for transport-I (ESCRT-I) complex, which is conserved from yeast to humans, directs the lysosomal degradation of ubiquitinated transmembrane proteins and the budding of the HIV virus. Yeast ESCRT-I contains four subunits, Vps23, Vps28, Vps37, and Mvb12. The crystal structure of the heterotetrameric ESCRT-I complex reveals a highly asymmetric complex of 1:1:1:1 subunit stoichiometry. The core complex is nearly 18 nm long and consists of a headpiece attached to a 13 nm stalk. The stalk is important for cargo sorting by ESCRT-I and is proposed to serve as a spacer regulating the correct disposition of cargo and other ESCRT components. Hydrodynamic constraints and crystallographic structures were used to generate a model of intact ESCRT-I in solution. The results show how ESCRT-I uses a combination of a rigid stalk and flexible tethers to interact with lipids, cargo, and other ESCRT complexes over a span of approximately 25 nm.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, U.S. Department of Health and Human Services, Bethesda, MD 20892, USA.
Organizational Affiliation: