Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Fisher, R.D., Chung, H.Y., Zhai, Q., Robinson, H., Sundquist, W.I., Hill, C.P.(2007) Cell 128: 841-852
- PubMed: 17350572
- DOI: https://doi.org/10.1016/j.cell.2007.01.035
- Primary Citation of Related Structures:
2OEV, 2OEW, 2OEX - PubMed Abstract:
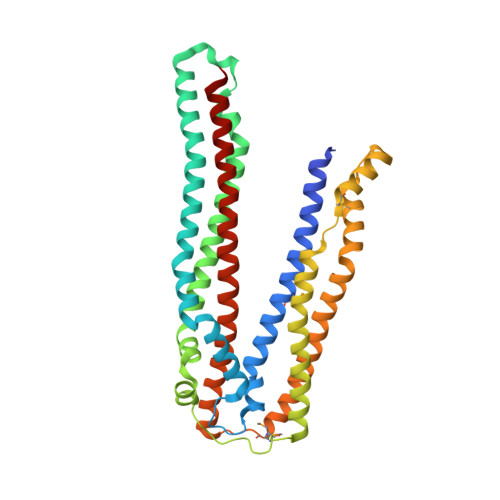
ALIX/AIP1 functions in enveloped virus budding, endosomal protein sorting, and many other cellular processes. Retroviruses, including HIV-1, SIV, and EIAV, bind and recruit ALIX through YPX(n)L late-domain motifs (X = any residue; n = 1-3). Crystal structures reveal that human ALIX is composed of an N-terminal Bro1 domain and a central domain that is composed of two extended three-helix bundles that form elongated arms that fold back into a "V." The structures also reveal conformational flexibility in the arms that suggests that the V domain may act as a flexible hinge in response to ligand binding. YPX(n)L late domains bind in a conserved hydrophobic pocket on the second arm near the apex of the V, whereas CHMP4/ESCRT-III proteins bind a conserved hydrophobic patch on the Bro1 domain, and both interactions are required for virus budding. ALIX therefore serves as a flexible, extended scaffold that connects retroviral Gag proteins to ESCRT-III and other cellular-budding machinery.
- Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT 84112, USA.
Organizational Affiliation: