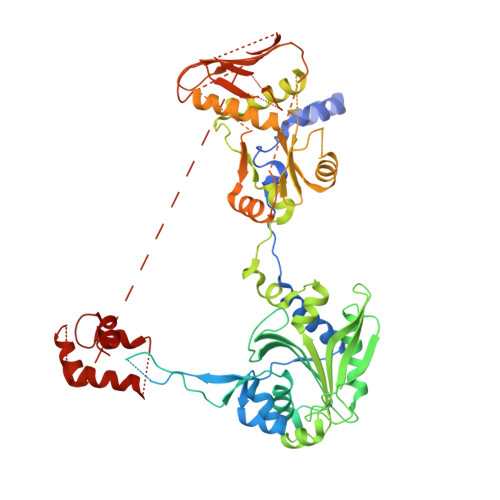
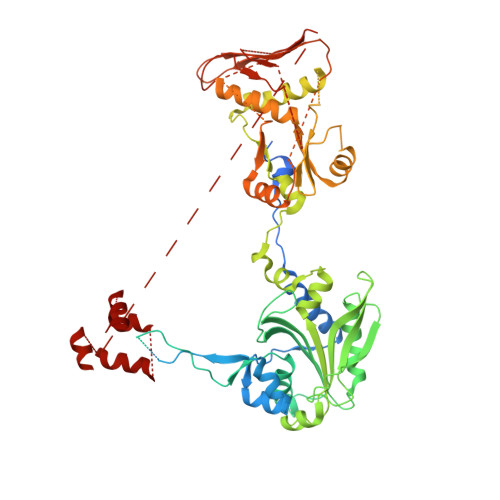
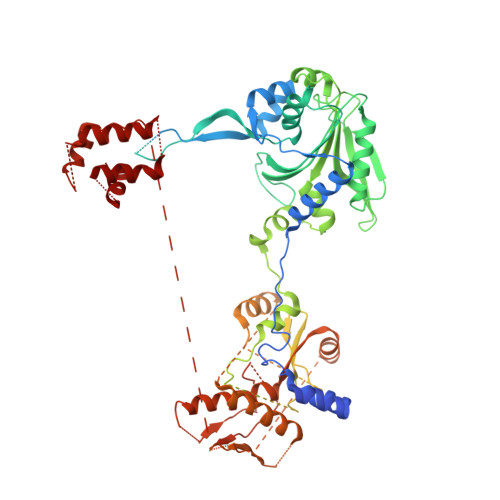
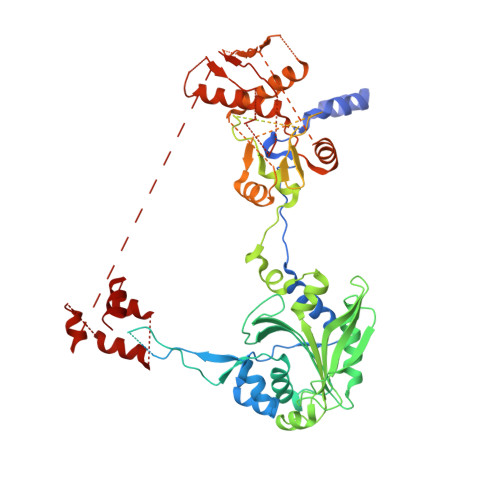
Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Kamtekar, S., Hohn, M.J., Park, H.S., Schnitzbauer, M., Sauerwald, A., Soll, D., Steitz, T.A.(2007) Proc Natl Acad Sci U S A 104: 2620-2625
- PubMed: 17301225
- DOI: https://doi.org/10.1073/pnas.0611504104
- Primary Citation of Related Structures:
2ODR - PubMed Abstract:
A number of archaeal organisms generate Cys-tRNA(Cys) in a two-step pathway, first charging phosphoserine (Sep) onto tRNA(Cys) and subsequently converting it to Cys-tRNA(Cys). We have determined, at 3.2-A resolution, the structure of the Methanococcus maripaludis phosphoseryl-tRNA synthetase (SepRS), which catalyzes the first step of this pathway. The structure shows that SepRS is a class II, alpha(4) synthetase whose quaternary structure arrangement of subunits closely resembles that of the heterotetrameric (alphabeta)(2) phenylalanyl-tRNA synthetase (PheRS). Homology modeling of a tRNA complex indicates that, in contrast to PheRS, a single monomer in the SepRS tetramer may recognize both the acceptor terminus and anticodon of a tRNA substrate. Using a complex with tungstate as a marker for the position of the phosphate moiety of Sep, we suggest that SepRS and PheRS bind their respective amino acid substrates in dissimilar orientations by using different residues.
- Department of Molecular Biophysics and Biochemistry and Chemistry and Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8114, USA.
Organizational Affiliation: