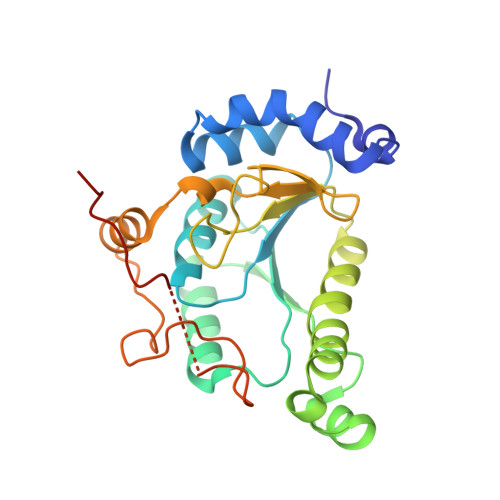
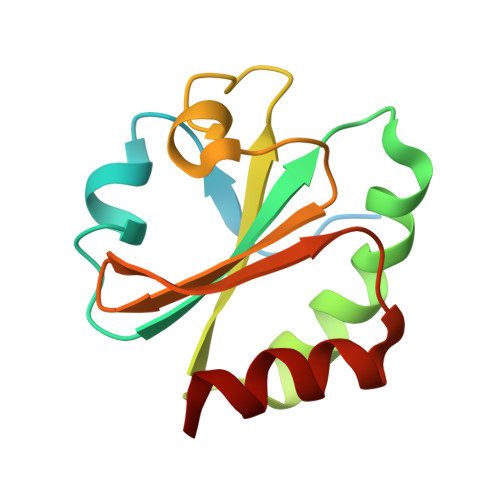
3'-Phosphoadenosine-5'-phosphosulfate Reductase in Complex with Thioredoxin: A Structural Snapshot in the Catalytic Cycle.
Chartron, J., Shiau, C., Stout, C.D., Carroll, K.S.(2007) Biochemistry 46: 3942-3951
- PubMed: 17352498
- DOI: https://doi.org/10.1021/bi700130e
- Primary Citation of Related Structures:
2O8V - PubMed Abstract:
The crystal structure of Escherichia coli 3'-phosphoadenosine-5'-phosphosulfate (PAPS) reductase in complex with E. coli thioredoxin 1 (Trx1) has been determined to 3.0 A resolution. The two proteins are covalently linked via a mixed disulfide that forms during nucleophilic attack of Trx's N-terminal cysteine on the Sgamma atom of the PAPS reductase S-sulfocysteine (E-Cys-Sgamma-SO3-), a central intermediate in the catalytic cycle. For the first time in a crystal structure, residues 235-244 in the PAPS reductase C-terminus are observed, depicting an array of interprotein salt bridges between Trx and the strictly conserved glutathione-like sequence, Glu238Cys239Gly240Leu241His242. The structure also reveals a Trx-binding surface adjacent to the active site cleft and regions of PAPS reductase associated with conformational change. Interaction at this site strategically positions Trx to bind the S-sulfated C-terminus and addresses the mechanism for requisite structural rearrangement of this domain. An apparent sulfite-binding pocket at the protein-protein interface explicitly orients the S-sulfocysteine Sgamma atom for nucleophilic attack in a subsequent step. Taken together, the structure of PAPS reductase in complex with Trx highlights the large structural rearrangement required to accomplish sulfonucleotide reduction and suggests a role for Trx in catalysis beyond the paradigm of disulfide reduction.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: