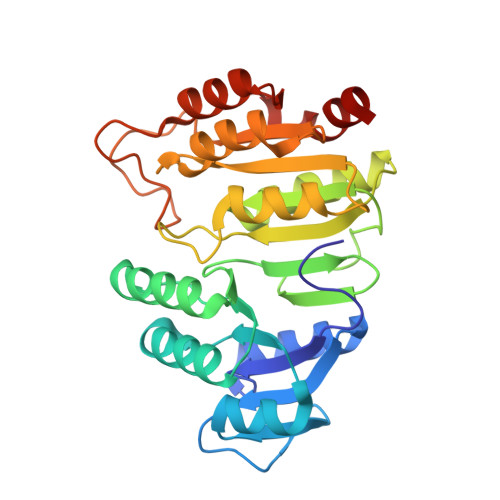
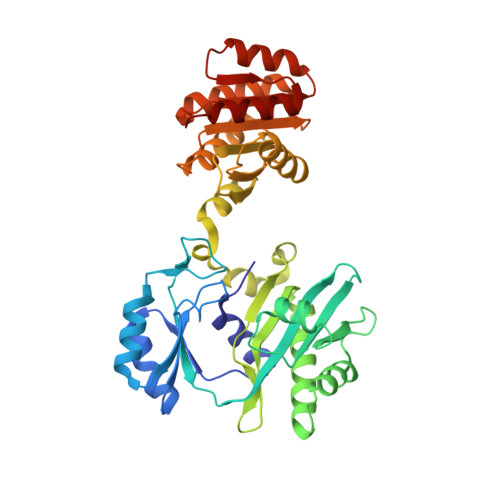
Participation of Cys 123alpha of Escherichia coli Succinyl-CoA Synthetase in Catalysis
Hidber, E., Brownie, E.R., Hayakawa, K., Fraser, M.E.(2007) Acta Crystallogr D Biol Crystallogr 63: 876-884
- PubMed: 17642514
- DOI: https://doi.org/10.1107/S0907444907029319
- Primary Citation of Related Structures:
2NU6, 2NU7, 2NU8, 2NU9, 2NUA - PubMed Abstract:
Succinyl-CoA synthetase has a highly conserved cysteine residue, Cys123alpha in the Escherichia coli enzyme, that is located near the CoA-binding site and the active-site histidine residue. To test whether the succinyl moiety of succinyl-CoA is transferred to the thiol of Cys123alpha as part of the catalytic mechanism, this residue was mutated to alanine, serine, threonine and valine. Each mutant protein was catalytically active, although less active than the wild type. This proved that the specific formation of a thioester bond with Cys123alpha is not part of the catalytic mechanism. To understand why the mutations affected catalysis, the crystal structures of the four mutant proteins were determined. The alanine mutant showed no structural changes yet had reduced activity, suggesting that the size of the cysteine is important for optimal activity. These results explain why this cysteine residue is conserved in the sequences of succinyl-CoA synthetases from different sources.
- Department of Biochemistry, University of Western Ontario, London, Ontario, Canada.
Organizational Affiliation: