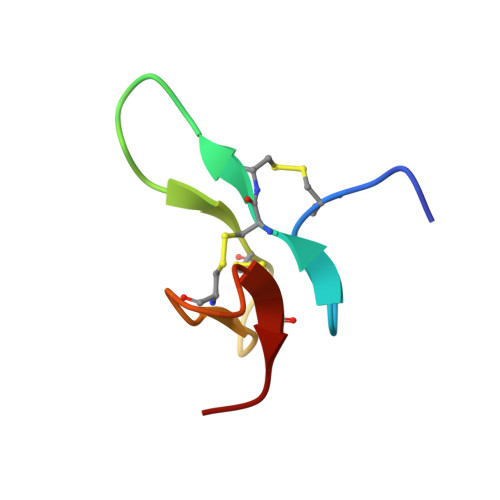
A Disulfide Stabilized beta-Sandwich Defines the Structure of a New Cysteine Framework M-Superfamily Conotoxin
Kancherla, A.K., Meesala, S., Jorwal, P., Palanisamy, R., Sikdar, S.K., Sarma, S.P.(2015) ACS Chem Biol 10: 1847-1860
- PubMed: 25961405
- DOI: https://doi.org/10.1021/acschembio.5b00226
- Primary Citation of Related Structures:
2MW7 - PubMed Abstract:
The structure of a new cysteine framework (-C-CC-C-C-C-) "M"-superfamily conotoxin, Mo3964, shows it to have a β-sandwich structure that is stabilized by inter-sheet cross disulfide bonds. Mo3964 decreases outward K(+) currents in rat dorsal root ganglion neurons and increases the reversal potential of the NaV1.2 channels. The structure of Mo3964 (PDB ID: 2MW7 ) is constructed from the disulfide connectivity pattern, i.e., 1-3, 2-5, and 4-6, that is hitherto undescribed for the "M"-superfamily conotoxins. The tertiary structural fold has not been described for any of the known conus peptides. NOE (549), dihedral angle (84), and hydrogen bond (28) restraints, obtained by measurement of (h3)JNC' scalar couplings, were used as input for structure calculation. The ensemble of structures showed a backbone root mean square deviation of 0.68 ± 0.18 Å, with 87% and 13% of the backbone dihedral (ϕ, ψ) angles lying in the most favored and additional allowed regions of the Ramachandran map. The conotoxin Mo3964 represents a new bioactive peptide fold that is stabilized by disulfide bonds and adds to the existing repertoire of scaffolds that can be used to design stable bioactive peptide molecules.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore-560012, India.
Organizational Affiliation: