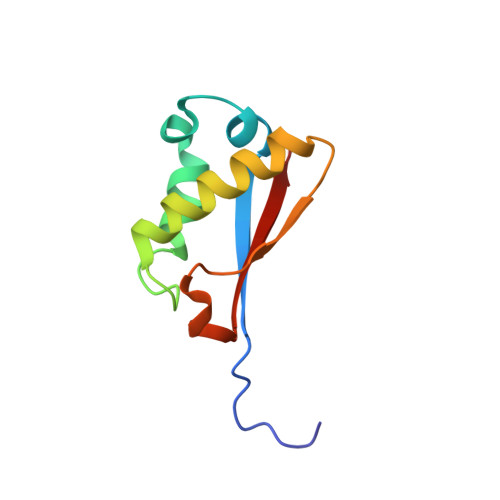
NMR structure and dynamics of Q4D059, a kinetoplastid-specific and conserved protein from Trypanosoma cruzi.
Lopez-Castilla, A., Pons, T., Pires, J.R.(2015) J Struct Biol 190: 11-20
- PubMed: 25748338
- DOI: https://doi.org/10.1016/j.jsb.2015.02.007
- Primary Citation of Related Structures:
2MNI - PubMed Abstract:
Q4D059 (UniProt accession number), is an 86-residue protein from Trypanosoma cruzi, conserved in the related kinetoplastid parasites Trypanosoma brucei and Leishmania major. These pathogens are the causal agents of the neglected diseases: Chagas, sleeping sickness and leishmaniases respectively and had recently their genomes sequenced. Q4D059 shows low sequence similarity with mammal proteins and because of its essentiality demonstrated in T. brucei, it is a potential target for anti-parasitic drugs. The 11 hypothetical proteins homologous to Q4D059 are all uncharacterized proteins of unknown function. Here, the solution structure of Q4D059 was solved by NMR and its backbone dynamics was characterized by (15)N relaxation parameters. The structure is composed by a parallel/anti-parallel three-stranded β-sheet packed against four helical regions. The structure is well defined by ca. 9 NOEs per residue and a backbone rmsd of 0.50±0.05 Å for the representative ensemble of 20 lowest-energy structures. The structure is overall rigid except for N-terminal residues A(9) to D(11) at the beginning of β1, K(38), V(39) at the end of helix H3 with rapid motion in the ps-ns timescale and G(25) (helix H2), I(68) (β2) and V(78) (loop 3) undergoing internal motion in the μs-ms timescale. Limited structural similarities were found in protein structures deposited in the PDB, therefore functional inferences based on protein structure information are not clear. Q4D059 adopts a α/β fold that is slightly similar to the ATPase sub-domain IIB of the heat-shock protein 70 (HSP70) and to the N-terminal domain of the ribosomal protein L11.
- Instituto de Bioquímica Médica Leopoldo de Meis, Universidade Federal do Rio de Janeiro, RJ, Brazil.
Organizational Affiliation: