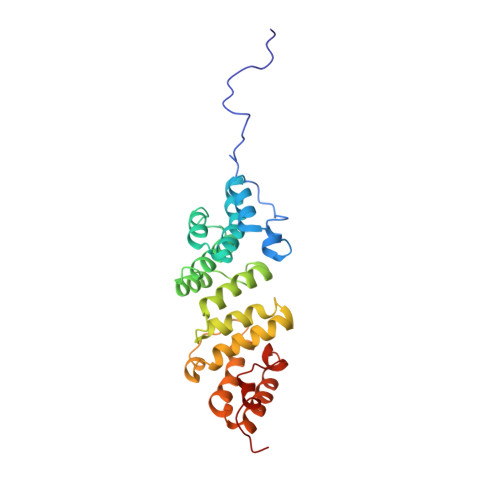
Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Jean, N.L., Bougault, C.M., Lodge, A., Derouaux, A., Callens, G., Egan, A.J., Ayala, I., Lewis, R.J., Vollmer, W., Simorre, J.P.(2014) Structure 22: 1047-1054
- PubMed: 24954617
- DOI: https://doi.org/10.1016/j.str.2014.04.017
- Primary Citation of Related Structures:
2MHK - PubMed Abstract:
The bacterial cell envelope contains the stress-bearing peptidoglycan layer, which is enlarged during cell growth and division by membrane-anchored synthases guided by cytoskeletal elements. In Escherichia coli, the major peptidoglycan synthase PBP1A requires stimulation by the outer-membrane-anchored lipoprotein LpoA. Whereas the C-terminal domain of LpoA interacts with PBP1A to stimulate its peptide crosslinking activity, little is known about the role of the N-terminal domain. Herein we report its NMR structure, which adopts an all-α-helical fold comprising a series of helix-turn-helix tetratricopeptide-repeat (TPR)-like motifs. NMR spectroscopy of full-length LpoA revealed two extended flexible regions in the C-terminal domain and limited, if any, flexibility between the N- and C-terminal domains. Analytical ultracentrifugation and small-angle X-ray scattering results are consistent with LpoA adopting an elongated shape, with dimensions sufficient to span from the outer membrane through the periplasm to interact with the peptidoglycan synthase PBP1A.
- University Grenoble Alpes, Institut de Biologie Structurale, F-38027 Grenoble, France; CEA, DSV, Institut de Biologie Structurale, F-38027 Grenoble, France; CNRS, Institut de Biologie Structurale, F-38027 Grenoble, France.
Organizational Affiliation: