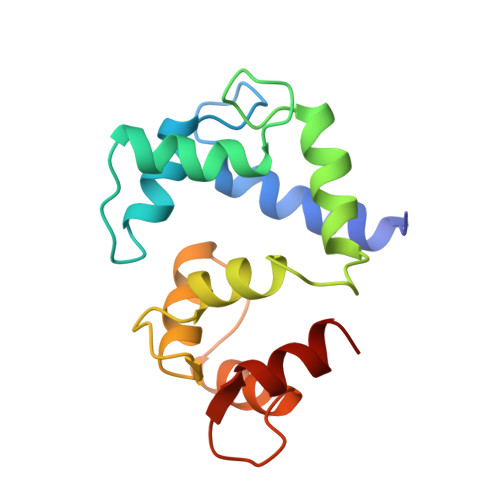
Capping of the N-terminus of PSD-95 by calmodulin triggers its postsynaptic release.
Zhang, Y., Matt, L., Patriarchi, T., Malik, Z.A., Chowdhury, D., Park, D.K., Renieri, A., Ames, J.B., Hell, J.W.(2014) EMBO J 33: 1341-1353
- PubMed: 24705785
- DOI: https://doi.org/10.1002/embj.201488126
- Primary Citation of Related Structures:
2MES - PubMed Abstract:
Postsynaptic density protein-95 (PSD-95) is a central element of the postsynaptic architecture of glutamatergic synapses. PSD-95 mediates postsynaptic localization of AMPA receptors and NMDA receptors and plays an important role in synaptic plasticity. PSD-95 is released from postsynaptic membranes in response to Ca(2+) influx via NMDA receptors. Here, we show that Ca(2+)/calmodulin (CaM) binds at the N-terminus of PSD-95. Our NMR structure reveals that both lobes of CaM collapse onto a helical structure of PSD-95 formed at its N-terminus (residues 1-16). This N-terminal capping of PSD-95 by CaM blocks palmitoylation of C3 and C5, which is required for postsynaptic PSD-95 targeting and the binding of CDKL5, a kinase important for synapse stability. CaM forms extensive hydrophobic contacts with Y12 of PSD-95. The PSD-95 mutant Y12E strongly impairs binding to CaM and Ca(2+)-induced release of PSD-95 from the postsynaptic membrane in dendritic spines. Our data indicate that CaM binding to PSD-95 serves to block palmitoylation of PSD-95, which in turn promotes Ca(2+)-induced dissociation of PSD-95 from the postsynaptic membrane.
- Department of Chemistry, University of California, Davis, CA, USA.
Organizational Affiliation: