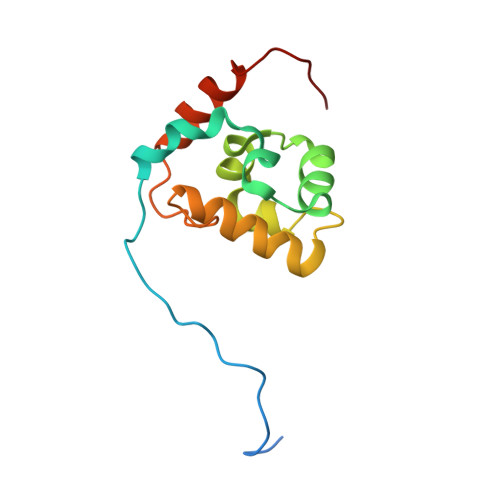
Helicobacter pylori RNA polymerase alpha-subunit C-terminal domain shows features unique to -proteobacteria and binds NikR/DNA complexes.
Borin, B.N., Tang, W., Krezel, A.M.(2014) Protein Sci 23: 454-463
- PubMed: 24442709
- DOI: https://doi.org/10.1002/pro.2427
- Primary Citation of Related Structures:
2MAX - PubMed Abstract:
Bacterial RNA polymerase is a large, multi-subunit enzyme responsible for transcription of genomic information. The C-terminal domain of the α subunit of RNA polymerase (αCTD) functions as a DNA and protein recognition element localizing the polymerase on certain promoter sequences and is essential in all bacteria. Although αCTD is part of RNA polymerase, it is thought to have once been a separate transcription factor, and its primary role is the recruitment of RNA polymerase to various promoters. Despite the conservation of the subunits of RNA polymerase among bacteria, the mechanisms of regulation of transcription vary significantly. We have determined the tertiary structure of Helicobacter pylori αCTD. It is larger than other structurally determined αCTDs due to an extra, highly amphipathic helix near the C-terminal end. Residues within this helix are highly conserved among ɛ-proteobacteria. The surface of the domain that binds A/T rich DNA sequences is conserved and showed binding to DNA similar to αCTDs of other bacteria. Using several NikR dependent promoter sequences, we observed cooperative binding of H. pylori αCTD to NikR:DNA complexes. We also produced αCTD lacking the 19 C-terminal residues, which showed greatly decreased stability, but maintained the core domain structure and binding affinity to NikR:DNA at low temperatures. The modeling of H. pylori αCTD into the context of transcriptional complexes suggests that the additional amphipathic helix mediates interactions with transcriptional regulators.
- Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee, 37232.
Organizational Affiliation: