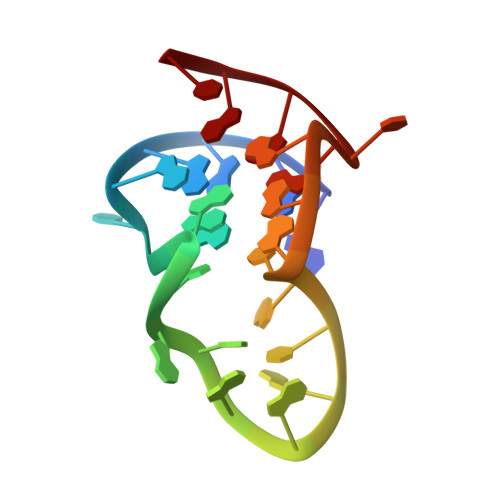
Formation of Pearl-Necklace Monomorphic G-Quadruplexes in the Human CEB25 Minisatellite.
Amrane, S., Adrian, M., Heddi, B., Serero, A., Nicolas, A., Mergny, J.L., Phan, A.T.(2012) J Am Chem Soc 134: 5807-5816
- PubMed: 22376028
- DOI: https://doi.org/10.1021/ja208993r
- Primary Citation of Related Structures:
2LPW - PubMed Abstract:
CEB25 is a human minisatellite locus, composed of slightly polymorphic 52-nucleotide (nt) tandem repeats. Genetically, most if not all individuals of the human population are heterozygous, carrying alleles ranging from 0.5 to 20 kb, maintained by mendelian inheritance but also subject to germline instability. To provide insights on the biological role of CEB25, we interrogated its structural features. We report the NMR structure of the G-quadruplex formed by the conserved 26-nt G-rich fragment of the CEB25 motif. In K(+) solution, this sequence forms a propeller-type parallel-stranded G-quadruplex involving a 9-nt central double-chain-reversal loop. This long loop is anchored to the 5'-end of the sequence by an A·T Watson-Crick base pair and a potential G·A noncanonical base pair. These base pairs contribute to the stability of the overall G-quadruplexstructure, as measured by an increase of about 17 kcal/mol in enthalpy or 6 °C in melting temperature. Further, we demonstrate that such a monomorphic structure is formed within longer sequence contexts folding into a pearl-necklace structure.
- School of Physical and Mathematical Sciences, Nanyang Technological University, 637371 Singapore.
Organizational Affiliation: