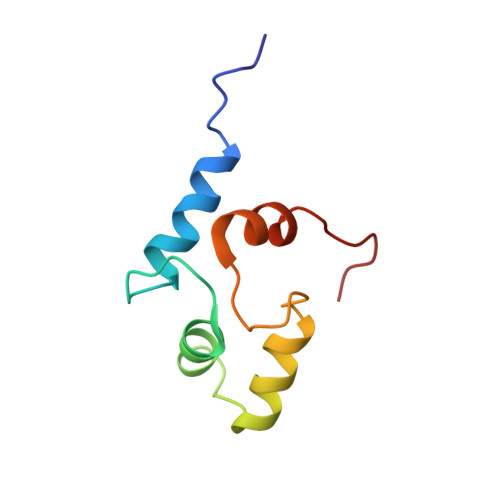
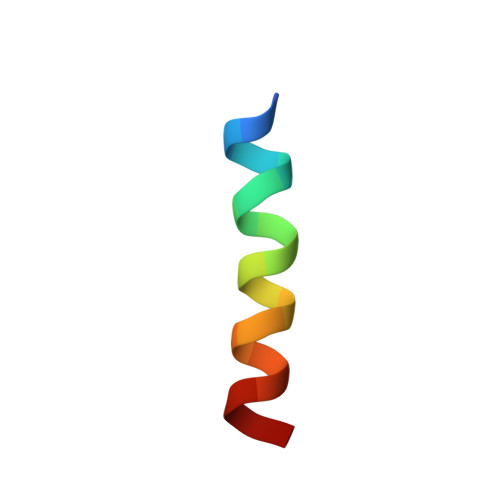Structural basis for Ca2+-induced activation and dimerization of estrogen receptor alpha by calmodulin.
Zhang, Y., Li, Z., Sacks, D.B., Ames, J.B.(2012) J Biological Chem 287: 9336-9344
- PubMed: 22275375
- DOI: https://doi.org/10.1074/jbc.M111.334797
- Primary Citation of Related Structures:
2LLO, 2LLQ - PubMed Abstract:
The estrogen receptor α (ER-α) regulates expression of target genes implicated in development, metabolism, and breast cancer. Calcium-dependent regulation of ER-α is critical for activating gene expression and is controlled by calmodulin (CaM). Here, we present the NMR structures for the two lobes of CaM each bound to a localized region of ER-α (residues 287-305). A model of the complete CaM·ER-α complex was constructed by combining these two structures with additional data. The two lobes of CaM both compete for binding at the same site on ER-α (residues 292, 296, 299, 302, and 303), which explains why full-length CaM binds two molecules of ER-α in a 1:2 complex and stabilizes ER-α dimerization. Exposed glutamate residues in CaM (Glu(11), Glu(14), Glu(84), and Glu(87)) form salt bridges with key lysine residues in ER-α (Lys(299), Lys(302), and Lys(303)), which are likely to prevent ubiquitination at these sites and inhibit degradation of ER-α. Mutants of ER-α at the CaM-binding site (W292A and K299A) weaken binding to CaM, and I298E/K299D disrupts estrogen-induced transcription. CaM facilitates dimerization of ER-α in the absence of estrogen, and stimulation of ER-α by either Ca(2+) and/or estrogen may serve to regulate transcription in a combinatorial fashion.
- Department of Chemistry, University of California, Davis, California 95616, USA.
Organizational Affiliation: