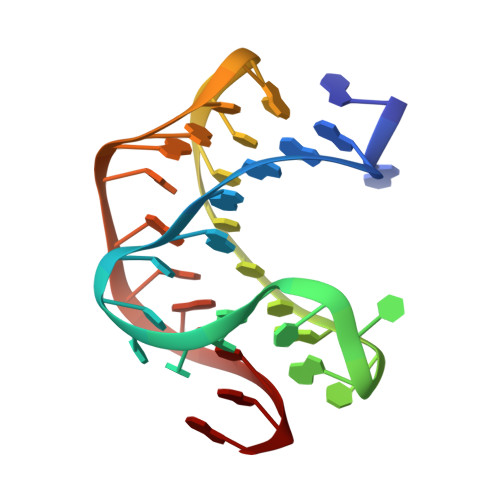
Structural Insights into Riboswitch Control of the Biosynthesis of Queuosine, a Modified Nucleotide Found in the Anticodon of tRNA
Kang, M., Peterson, R.D., Feigon, J.(2009) Mol Cell 33: 784-790
- PubMed: 19285444
- DOI: https://doi.org/10.1016/j.molcel.2009.02.019
- Primary Citation of Related Structures:
2L1V - PubMed Abstract:
The modified nucleotide queuosine (Q) is almost universally found in the anticodon wobble position of specific tRNAs. In many bacteria, biosynthesis of Q is modulated by a class of regulatory mRNA elements called riboswitches. The preQ(1) riboswitch, found in the 5'UTR of bacterial genes involved in synthesis of the Q precursors preQ(0) and preQ(1), contains the smallest known aptamer domain. We report the solution structure of the preQ(1) riboswitch aptamer domain from Bacillus subtilis bound to preQ(1), which is a unique compact pseudoknot with three loops and two stems that encapsulates preQ(1) at the junction between the two stems. The pseudoknot only forms in the presence of preQ(1), and the 3' A-rich tail of the aptamer domain is an integral part of the pseudoknot. In the absence of preQ(1), the A-rich tail forms part of the antiterminator. These structural studies provide insight into riboswitch transcriptional control of preQ(1) biosynthesis.
- UCLA-DOE Institute for Genomics and Proteomics, University of California, Los Angeles, Los Angeles, CA 90095-1569, USA.
Organizational Affiliation: