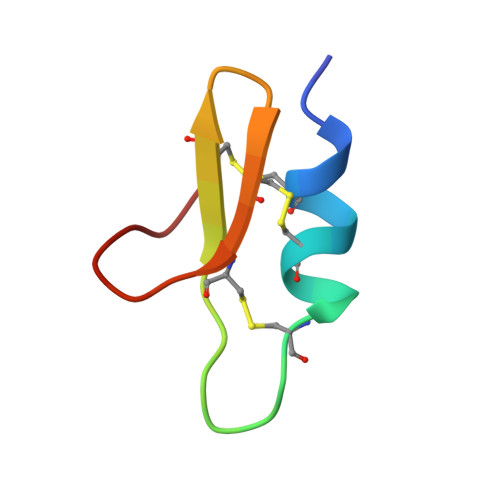
Solution structure of BTK-2, a novel hK(v)1.1 inhibiting scorpion toxin, from the eastern Indian scorpion Mesobuthus tamulus.
Kumar, G.S., Upadhyay, S., Mathew, M.K., Sarma, S.P.(2011) Biochim Biophys Acta 1814: 459-469
- PubMed: 21256986
- DOI: https://doi.org/10.1016/j.bbapap.2011.01.006
- Primary Citation of Related Structures:
2KTC - PubMed Abstract:
The three dimensional structure of a 32 residue three disulfide scorpion toxin, BTK-2, from the Indian red scorpion Mesobuthus tamulus has been determined using isotope edited solution NMR methods. Samples for structural and electrophysiological studies were prepared using recombinant DNA methods. Electrophysiological studies show that the peptide is active against hK(v)1.1 channels. The structure of BTK-2 was determined using 373 distance restraints from NOE data, 66 dihedral angle restraints from NOE, chemical shift and scalar coupling data, 6 constraints based on disulfide linkages and 8 constraints based on hydrogen bonds. The root mean square deviation (r.m.s.d) about the averaged co-ordinates of the backbone (N, C(α), C') and all heavy atoms are 0.81 ± 0.23Å and 1.51 ± 0.29Å respectively. The backbone dihedral angles (ϕ and ψ) for all residues occupy the favorable and allowed regions of the Ramachandran map. The three dimensional structure of BTK-2 is composed of three well defined secondary structural regions that constitute the α-β-β structural motif. Comparisons between the structure of BTK-2 and other closely related scorpion toxins pointed towards distinct differences in surface properties that provide insights into the structure-function relationships among this important class of voltage-gated potassium channel inhibiting peptides.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.
Organizational Affiliation: