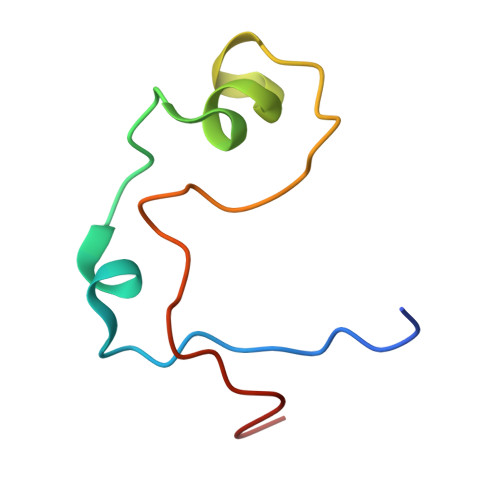
Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia.
Cierpicki, T., Risner, L.E., Grembecka, J., Lukasik, S.M., Popovic, R., Omonkowska, M., Shultis, D.D., Zeleznik-Le, N.J., Bushweller, J.H.(2010) Nat Struct Mol Biol 17: 62-68
- PubMed: 20010842
- DOI: https://doi.org/10.1038/nsmb.1714
- Primary Citation of Related Structures:
2KKF - PubMed Abstract:
The gene MLL (encoding the protein mixed-lineage leukemia) is the target of chromosomal translocations that cause leukemias with poor prognosis. All leukemogenic MLL fusion proteins retain the CXXC domain, which binds to nonmethylated CpG DNA sites. We present the solution structure of the MLL CXXC domain in complex with DNA, showing how the CXXC domain distinguishes nonmethylated from methylated CpG DNA. On the basis of the structure, we generated point mutations that disrupt DNA binding. Introduction of these mutations into the MLL-AF9 fusion protein resulted in increased DNA methylation of specific CpG nucleotides in Hoxa9, increased H3K9 methylation, decreased expression of Hoxa9-locus transcripts, loss of immortalization potential, and inability to induce leukemia in mice. These results establish that DNA binding by the CXXC domain and protection against DNA methylation is essential for MLL fusion leukemia. They also provide support for viewing this interaction as a potential target for therapeutic intervention.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, Virginia, USA.
Organizational Affiliation: