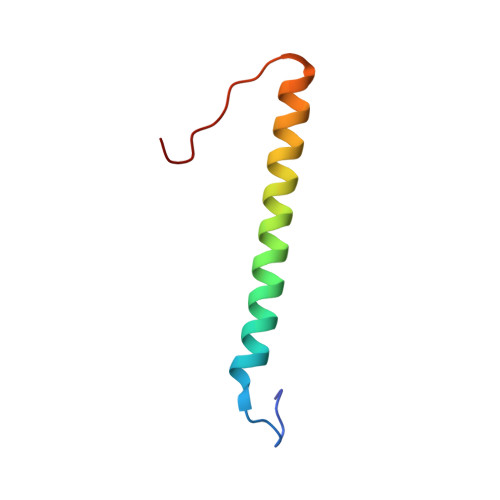
Solution structure, determined by nuclear magnetic resonance, of the b30-82 domain of subunit b of Escherichia coli F1Fo ATP synthase
Priya, R., Biukovic, G., Gayen, S., Vivekanandan, S., Gruber, G.(2009) J Bacteriol 191: 7538-7544
- PubMed: 19820091
- DOI: https://doi.org/10.1128/JB.00540-09
- Primary Citation of Related Structures:
2KHK - PubMed Abstract:
Subunit b, the peripheral stalk of bacterial F(1)F(o) ATP synthases, is composed of a membrane-spanning and a soluble part. The soluble part is divided into tether, dimerization, and delta-binding domains. The first solution structure of b30-82, including the tether region and part of the dimerization domain, has been solved by nuclear magnetic resonance, revealing an alpha-helix between residues 39 and 72. In the solution structure, b30-82 has a length of 48.07 A. The surface charge distribution of b30-82 shows one side with a hydrophobic surface pattern, formed by alanine residues. Alanine residues 61, 68, 70, and 72 were replaced by single cysteines in the soluble part of subunit b, b22-156. The cysteines at positions 61, 68, and 72 showed disulfide formation. In contrast, no cross-link could be formed for the A70C mutant. The patterns of disulfide bonding, together with the circular dichroism spectroscopy data, are indicative of an adjacent arrangement of residues 61, 68, and 72 in both alpha-helices in b22-156.
- School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore.
Organizational Affiliation: