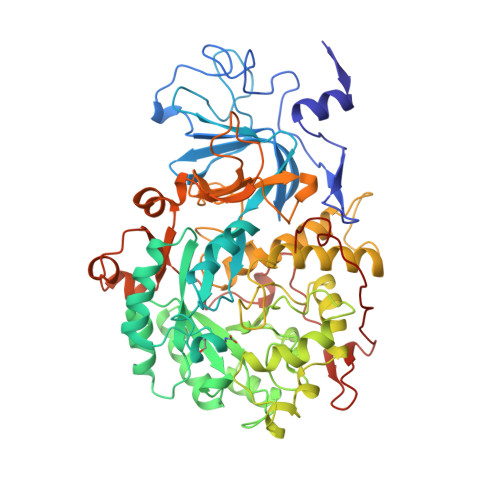
The crystal structure of urease from Klebsiella aerogenes.
Jabri, E., Carr, M.B., Hausinger, R.P., Karplus, P.A.(1995) Science 268: 998-1004
- PubMed: 7754395
- Primary Citation of Related Structures:
2KAU - PubMed Abstract:
The crystal structure of urease from Klebsiella aerogenes has been determined at 2.2 A resolution and refined to an R factor of 18.2 percent. The enzyme contains four structural domains: three with novel folds playing structural roles, and an (alpha beta)8 barrel domain, which contains the bi-nickel center. The two active site nickels are 3.5 A apart. One nickel ion is coordinated by three ligands (with low occupancy of a fourth ligand) and the second is coordinated by five ligands. A carbamylated lysine provides an oxygen ligand to each nickel, explaining why carbon dioxide is required for the activation of urease apoenzyme. The structure is compatible with a catalytic mechanism whereby urea ligates Ni-1 to complete its tetrahedral coordination and a hydroxide ligand of Ni-2 attacks the carbonyl carbon. A surprisingly high structural similarity between the urease catalytic domain and that of the zinc-dependent adenosine deaminase reveals a remarkable example of active site divergence.
- Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: