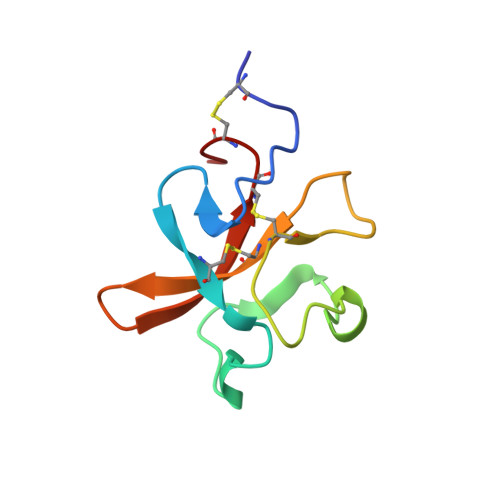
NMR Solution Structure of the Neurotrypsin Kringle Domain
Ozhogina, O.A., Grishaev, A., Bominaar, E.L., Llinas, M.(2008) Biochemistry 47: 12290-12298
- PubMed: 18956887
- DOI: https://doi.org/10.1021/bi800555z
- Primary Citation of Related Structures:
2K4R, 2K51 - PubMed Abstract:
Neurotrypsin is a multidomain protein that serves as a brain-specific serine protease. Here we report the NMR structure of its kringle domain, NT/K. The data analysis was performed with the BACUS (Bayesian analysis of coupled unassigned spins) algorithm. This study presents the first application of BACUS to the structure determination of a 13C unenriched protein for which no prior experimental 3D structure was available. NT/K adopts the kringle fold, consisting of an antiparallel beta-sheet bridged by an overlapping pair of disulfides. The structure reveals the presence of a surface-exposed left-handed polyproline II helix that is closely packed to the core beta-structure. This feature distinguishes NT/K from other members of the kringle fold and points toward a novel functional role for a kringle domain. Functional divergence among kringle domains is discussed on the basis of their surface and electrostatic characteristics.
- Department of Chemistry, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, USA. oao.sputnik@gmail.com
Organizational Affiliation: