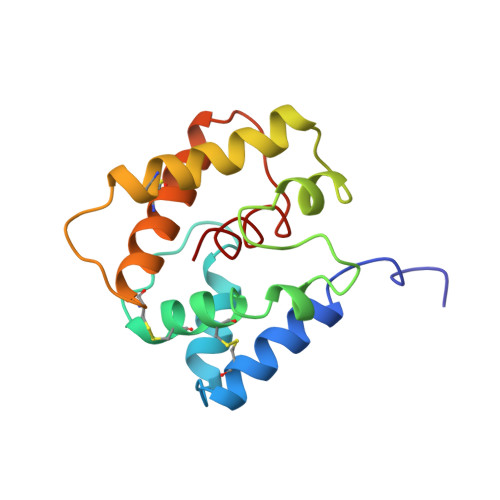
Structural Basis of Ligand Binding and Release in Insect Pheromone-Binding Proteins: NMR Structure of Antheraea polyphemus PBP1 at pH 4.5
Damberger, F.F., Ishida, Y., Leal, W.S., Wuthrich, K.(2007) J Mol Biology 373: 811-819
- PubMed: 17884092
- DOI: https://doi.org/10.1016/j.jmb.2007.07.078
- Primary Citation of Related Structures:
2JPO - PubMed Abstract:
The NMR structure of the Antheraea polyphemus pheromone-binding protein 1 at pH 4.5, ApolPBP1A, was determined at 20 degrees C. The structure consists of six alpha-helices, which are arranged in a globular fold that encapsulates a central helix alpha7 formed by the C-terminal polypeptide segment 131-142. The 3D arrangement of these helices is anchored by the three disulfide bonds 19-54, 50-108 and 97-117, which were identified by NMR. Superposition of the ApolPBP1A structure with the structure of the homologous pheromone-binding protein of Bombyx mori at pH 4.5, BmorPBPA, yielded an rmsd of 1.7 A calculated for the backbone heavy-atoms N, Calpha and C' of residues 10-142. In contrast, the present ApolPBP1A structure is different from a recently proposed molecular model for a low-pH form of ApolPBP1 that does not contain the central helix alpha7. ApolPBP1 exhibits a pH-dependent transition between two different globular conformations in slow exchange on the NMR chemical shift timescale similar to BmorPBP, suggesting that the two proteins use the same mechanism of ligand binding and ejection. The extensive sequence homology observed for pheromone-binding proteins from moth species further implies that the previously proposed mechanism of ligand ejection involving the insertion of a C-terminal helix into the pheromone-binding site is a general feature of pheromone signaling in moths.
- Institute for Molecular Biology and Biophysics, ETH Zurich, 8093 Zurich, Switzerland.
Organizational Affiliation: