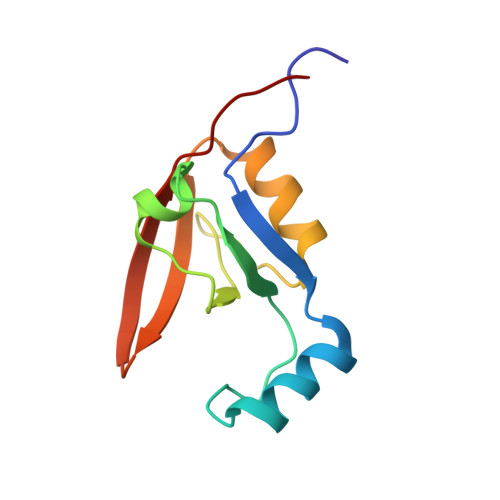
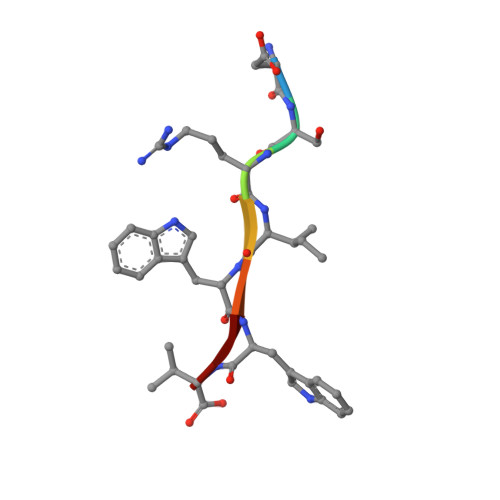
Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3
Runyon, S.T., Zhang, Y., Appleton, B.A., Sazinsky, S.L., Wu, P., Pan, B., Wiesmann, C., Skelton, N.J., Sidhu, S.S.(2007) Protein Sci 16: 2454-2471
- PubMed: 17962403
- DOI: https://doi.org/10.1110/ps.073049407
- Primary Citation of Related Structures:
2JOA, 2P3W - PubMed Abstract:
High-temperature requirement A (HtrA) and its homologs contain a serine protease domain followed by one or two PDZ domains. Bacterial HtrA proteins and the mitochondrial protein HtrA2/Omi maintain cell function by acting as both molecular chaperones and proteases to manage misfolded proteins. The biological roles of the mammalian family members HtrA1 and HtrA3 are less clear. We report a detailed structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3 using peptide libraries and affinity assays to define specificity, structural studies to view the molecular details of ligand recognition, and alanine scanning mutagenesis to investigate the energetic contributions of individual residues to ligand binding. In common with HtrA2/Omi, we show that the PDZ domains of HtrA1 and HtrA3 recognize hydrophobic polypeptides, and while C-terminal sequences are preferred, internal sequences are also recognized. However, the details of the interactions differ, as different domains rely on interactions with different residues within the ligand to achieve high affinity binding. The results suggest that mammalian HtrA PDZ domains interact with a broad range of hydrophobic binding partners. This promiscuous specificity resembles that of bacterial HtrA family members and suggests a similar function for recognizing misfolded polypeptides with exposed hydrophobic sequences. Our results support a common activation mechanism for the HtrA family, whereby hydrophobic peptides bind to the PDZ domain and induce conformational changes that activate the protease. Such a mechanism is well suited to proteases evolved for the recognition and degradation of misfolded proteins.
- Department of Medicinal Chemistry, Genetech, Inc., South San Francisco, CA 94080, USA.
Organizational Affiliation: