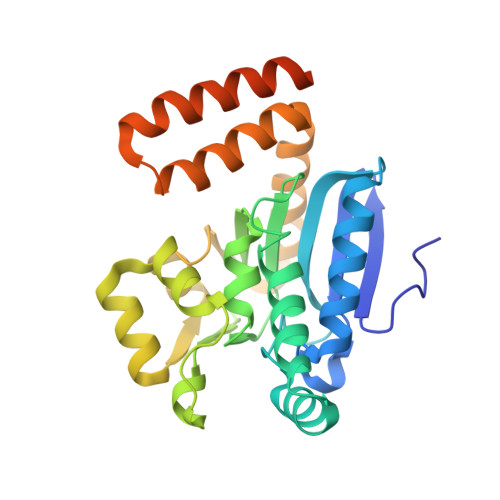
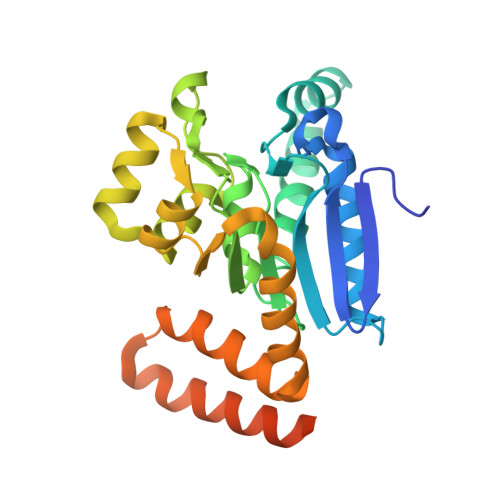
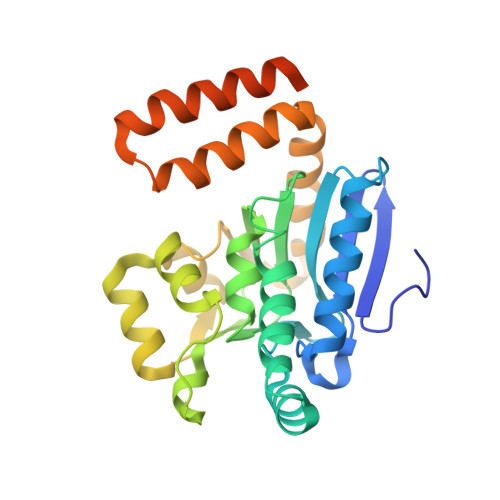
The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Leonard, P.M., Brzozowski, A.M., Lebedev, A., Marshall, C.M., Smith, D.J., Verma, C.S., Walton, N.J., Grogan, G.(2006) Acta Crystallogr D Biol Crystallogr 62: 1494
- PubMed: 17139085
- DOI: https://doi.org/10.1107/S0907444906039199
- Primary Citation of Related Structures:
2J5I - PubMed Abstract:
The crystal structure of hydroxycinnamoyl-CoA hydratase-lyase (HCHL) from Pseudomonas fluorescens AN103 has been solved to 1.8 A resolution. HCHL is a member of the crotonase superfamily and catalyses the hydration of the acyl-CoA thioester of ferulic acid [3-(4-hydroxy-3-methoxy-phenyl)prop-2-enoic acid] and the subsequent retro-aldol cleavage of the hydrated intermediate to yield vanillin (4-hydroxy-3-methoxy-benzaldehyde). The structure contains 12 molecules in the asymmetric unit, in which HCHL assumes a hexameric structure of two stacked trimers. The substrate, feruloyl-CoA, was modelled into the active site based on the structure of enoyl-CoA hydratase bound to the feruloyl-CoA-like substrate 4-(N,N-dimethylamino)-cinnamoyl-CoA (PDB code 1ey3). Feruloyl-CoA was bound in this model between helix 3 of the A subunit and helix 9 of the B subunit. A highly ordered structural water in the HCHL structure coincided with the thioester carbonyl of feruloyl-CoA in the model, suggesting that the oxyanion hole for stabilization of a thioester-derived enolate, characteristic of coenzyme-A dependent members of the crotonase superfamily, is conserved. The model also suggested that a strong hydrogen bond between the phenolic hydroxyl groups of feruloyl-CoA and BTyr239 may be an important determinant of the enzyme's ability to discriminate between the natural substrate and cinnamoyl-CoA, which is not a substrate.
- Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, England.
Organizational Affiliation: