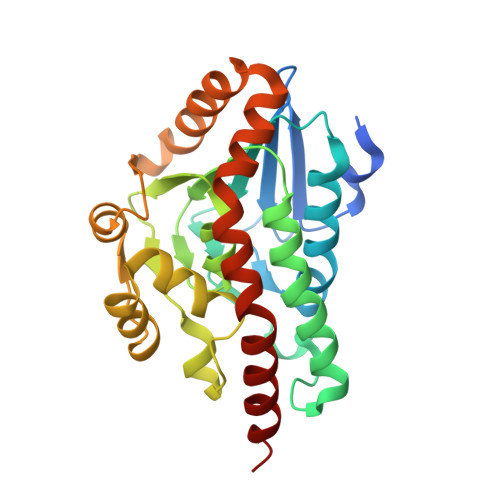
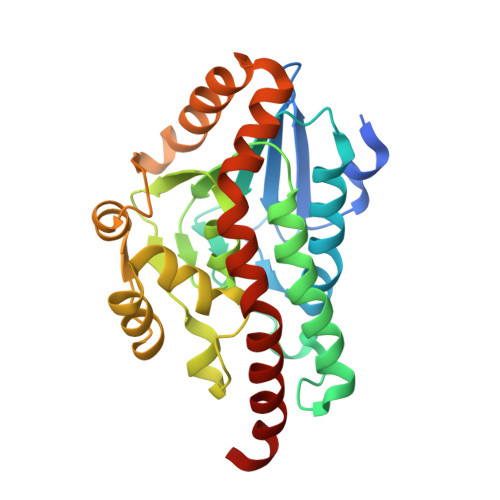
Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Bennett, J.P., Whittingham, J.L., Brzozowski, A.M., Leonard, P.M., Grogan, G.(2007) Biochemistry 46: 137
- PubMed: 17198383
- DOI: https://doi.org/10.1021/bi061900g
- Primary Citation of Related Structures:
2J5G, 2J5S - PubMed Abstract:
The gene alr4455 from the well-studied cyanobacterium Anabaena sp. PCC 7120 encodes a crotonase orthologue that displays beta-diketone hydrolase activity. Anabaena beta-diketone hydrolase (ABDH), in common with 6-oxocamphor hydrolase (OCH) from Rhodococcus sp. NCIMB 9784, catalyzes the desymmetrization of bicyclo[2.2.2]octane-2,6-dione to yield [(S)-3-oxocyclohexyl]acetic acid, a reaction unusual among the crotonase superfamily as the substrate is not an acyl-CoA thioester. The structure of ABDH has been determined to a resolution of 1.5 A in both native and ligand-bound forms. ABDH forms a hexamer similar to OCH and features one active site per enzyme monomer. The arrangement of side chains in the active site indicates that while the catalytic chemistry may be conserved in OCH orthologues, the structural determinants of substrate specificity are different. In the active site of ligand-bound forms that had been cocrystallized with the bicyclic diketone substrate bicyclo[2.2.2]octane-2,6-dione was found the product of the asymmetric enzymatic retro-Claisen reaction [(S)-3-oxocyclohexyl]acetic acid. The structures of ABDH in both native and ligand-bound forms reveal further details about structural variation and modes of coenzyme A-independent activity within the crotonases and provide further evidence of a wider suprafamily of enzymes that have recruited the crotonase fold for the catalysis of reactions other than those regularly attributed to canonical superfamily members.
- York Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, UK.
Organizational Affiliation: