The crystal structure of a hypothetical protein (Y1212_ARCFU) from Archeoglobus fulgidus
Tyagi, R., Burley, S.K., Swaminathan, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hypothetical protein AF_1212 | 139 | Archaeoglobus fulgidus | Mutation(s): 4 Gene Names: AF_1212 | 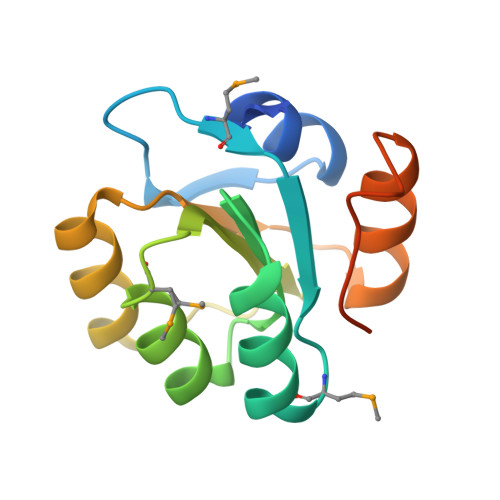 | |
UniProt | |||||
Find proteins for O29056 (Archaeoglobus fulgidus (strain ATCC 49558 / DSM 4304 / JCM 9628 / NBRC 100126 / VC-16)) Explore O29056 Go to UniProtKB: O29056 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O29056 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.409 | α = 90 |
| b = 66.016 | β = 90 |
| c = 122.392 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| CBASS | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SHELX | phasing |
| SHARP | phasing |