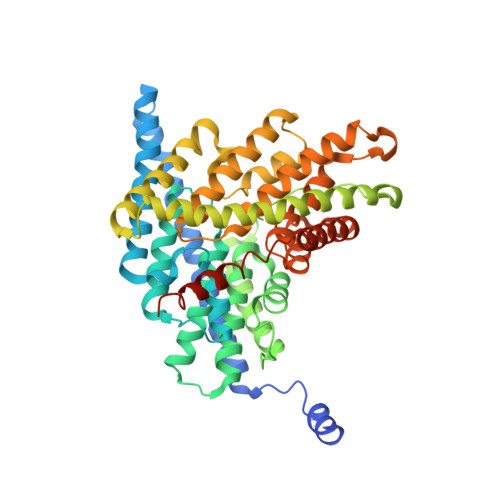
Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
Accardi, A., Lobet, S., Williams, C., Miller, C., Dutzler, R.(2006) J Mol Biology 362: 691-699
- PubMed: 16949616
- DOI: https://doi.org/10.1016/j.jmb.2006.07.081
- Primary Citation of Related Structures:
2HLF, 2HT2, 2HT3, 2HT4, 2HTK, 2HTL - PubMed Abstract:
The Cl-/H+ exchange-transporter CLC-ec1 mediates stoichiometric transmembrane exchange of two Cl- ions for one proton. A conserved tyrosine residue, Y445, coordinates one of the bound Cl- ions visible in the structure of this protein and is located near the intersection of the Cl- and H+ pathways. Mutants of this tyrosine were scrutinized for effects on the coupled transport of Cl- and H+ determined electrophysiologically and on protein structure determined crystallographically. Despite the strong conservation of Y445 in the CLC family, substitution of F or W at this position preserves wild-type transport behavior. Substitution by A, E, or H, however, produces uncoupled proteins with robust Cl- transport but greatly impaired movement of H+. The obligatory 2 Cl-/1 H+ stoichiometry is thus lost in these mutants. The structures of all the mutants are essentially identical to wild-type, but apparent anion occupancy in the Cl- binding region correlates with functional H+ coupling. In particular, as determined by anomalous diffraction in crystals grown in Br-, an electrophysiologically competent Cl- analogue, the well-coupled transporters show strong Br- electron density at the "inner" and "central" Cl- binding sites. However, in the uncoupled mutants, Br- density is absent at the central site, while still present at the inner site. An additional mutant, Y445L, is intermediate in both functional and structural features. This mutant clearly exchanges H+ for Cl-, but at a reduced H+-to-Cl- ratio; likewise, both the central and inner sites are occupied by Br-, but the central site shows lower Br- density than in wild-type (or in Y445F,W). The correlation between proton coupling and central-site occupancy argues that halide binding to the central transport site somehow facilitates movement of H+, a synergism that is not readily understood in terms of alternating-site antiport schemes.
- Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, MA 02454-9110, USA.
Organizational Affiliation: