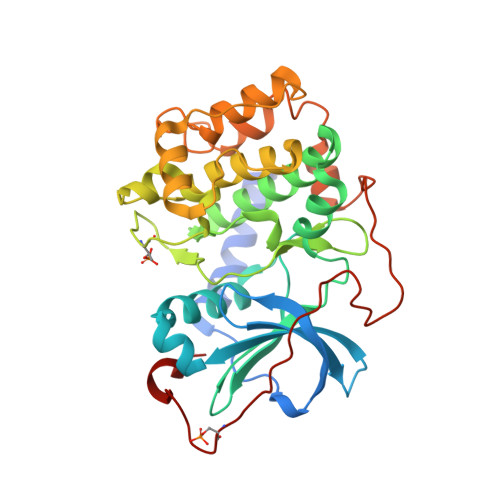
Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
Bonn, S., Herrero, S., Breitenlechner, C.B., Erlbruch, A., Lehmann, W., Engh, R.A., Gassel, M., Bossemeyer, D.(2006) J Biological Chem 281: 24818-24830
- PubMed: 16699172
- DOI: https://doi.org/10.1074/jbc.M512374200
- Primary Citation of Related Structures:
2GFC, 2GNF, 2GNG, 2GNH, 2GNI, 2GNJ, 2GNL - PubMed Abstract:
Controlling aberrant kinase-mediated cellular signaling is a major strategy in cancer therapy; successful protein kinase inhibitors such as Tarceva and Gleevec verify this approach. Specificity of inhibitors for the targeted kinase(s), however, is a crucial factor for therapeutic success. Based on homology modeling, we previously identified four amino acids in the active site of Rho-kinase that likely determine inhibitor specificities observed for Rho-kinase relative to protein kinase A (PKA) (in PKA numbering: T183A, L49I, V123M, and E127D), and a fifth (Q181K) that played a surprising role in PKA-PKB hybrid proteins. We have systematically mutated these residues in PKA to their counterparts in Rho-kinase, individually and in combination. Using four Rho-kinase-specific, one PKA-specific, and one pan-kinase-specific inhibitor, we measured the inhibitor-binding properties of the mutated proteins and identify the roles of individual residues as specificity determinants. Two combined mutant proteins, containing the combination of mutations T183A and L49I, closely mimic Rho-kinase. Kinetic results corroborate the hypothesis that side-chain identities form the major determinants of selectivity. An unexpected result of the analysis is the consistent contribution of the individual mutations by simple factors. Crystal structures of the surrogate kinase inhibitor complexes provide a detailed basis for an understanding of these selectivity determinant residues. The ability to obtain kinetic and structural data from these PKA mutants, combined with their Rho-kinase-like selectivity profiles, make them valuable for use as surrogate kinases for structure-based inhibitor design.
- Group of Structural Biochemistry, German Cancer Research Center, 69120 Heidelberg.
Organizational Affiliation: