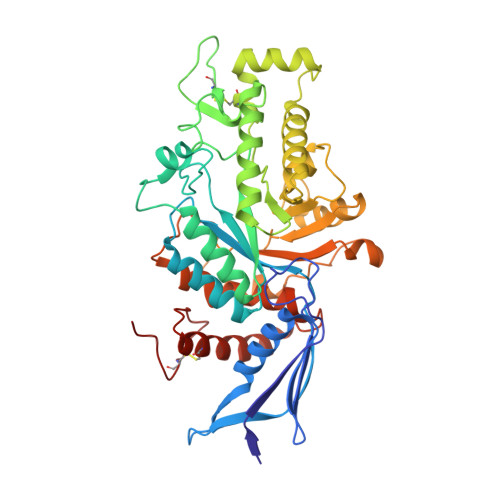
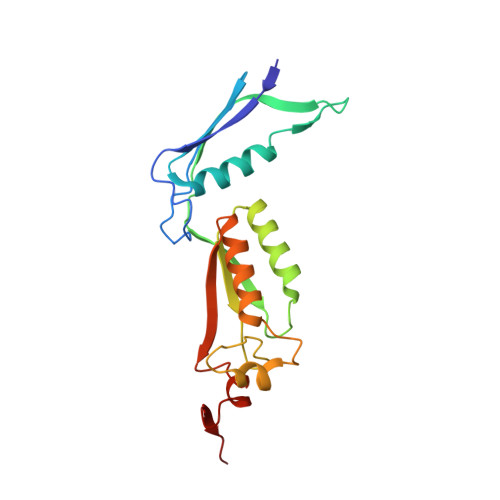
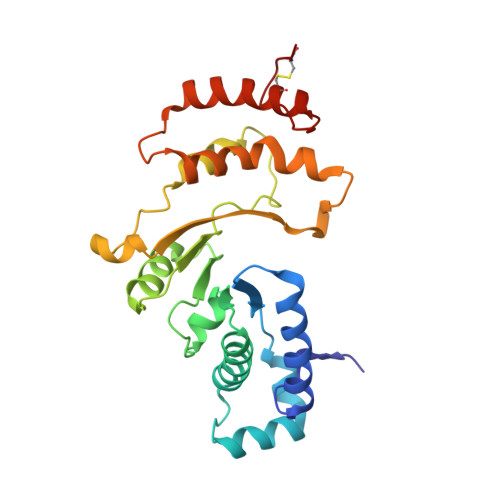
Crystallographic Structure of Human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of G(M2) Ganglioside Hydrolysis.
Lemieux, M.J., Mark, B.L., Cherney, M.M., Withers, S.G., Mahuran, D.J., James, M.N.(2006) J Mol Biology 359: 913-929
- PubMed: 16698036
- DOI: https://doi.org/10.1016/j.jmb.2006.04.004
- Primary Citation of Related Structures:
2GJX, 2GK1 - PubMed Abstract:
Lysosomal beta-hexosaminidase A (Hex A) is essential for the degradation of GM2 gangliosides in the central and peripheral nervous system. Accumulation of GM2 leads to severely debilitating neurodegeneration associated with Tay-Sachs disease (TSD), Sandoff disease (SD) and AB variant. Here, we present the X-ray crystallographic structure of Hex A to 2.8 A resolution and the structure of Hex A in complex with NAG-thiazoline, (NGT) to 3.25 A resolution. NGT, a mechanism-based inhibitor, has been shown to act as a chemical chaperone that, to some extent, prevents misfolding of a Hex A mutant associated with adult onset Tay Sachs disease and, as a result, increases the residual activity of Hex A to a level above the critical threshold for disease. The crystal structure of Hex A reveals an alphabeta heterodimer, with each subunit having a functional active site. Only the alpha-subunit active site can hydrolyze GM2 gangliosides due to a flexible loop structure that is removed post-translationally from beta, and to the presence of alphaAsn423 and alphaArg424. The loop structure is involved in binding the GM2 activator protein, while alphaArg424 is critical for binding the carboxylate group of the N-acetyl-neuraminic acid residue of GM2. The beta-subunit lacks these key residues and has betaAsp452 and betaLeu453 in their place; the beta-subunit therefore cleaves only neutral substrates efficiently. Mutations in the alpha-subunit, associated with TSD, and those in the beta-subunit, associated with SD are discussed. The effect of NGT binding in the active site of a mutant Hex A and its effect on protein function is discussed.
- CIHR Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alta., Canada T6G 2H7.
Organizational Affiliation: