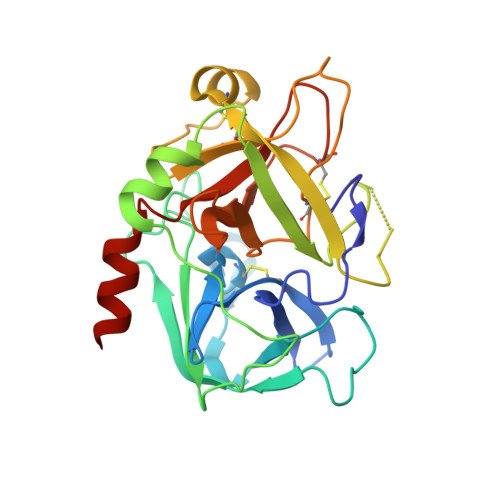
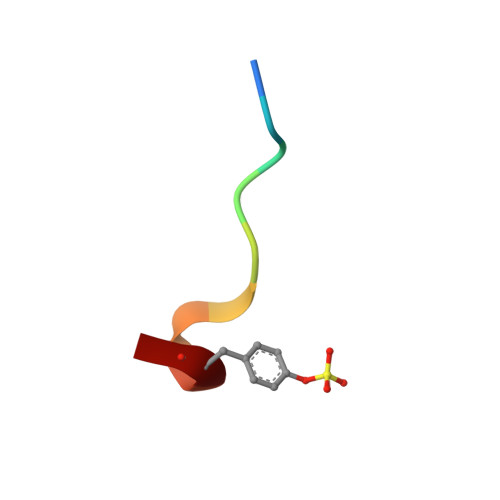
Total synthesis and structural confirmation of chlorodysinosin A.
Hanessian, S., Del Valle, J.R., Xue, Y., Blomberg, N.(2006) J Am Chem Soc 128: 10491-10495
- PubMed: 16895415
- DOI: https://doi.org/10.1021/ja0625834
- Primary Citation of Related Structures:
2GDE - PubMed Abstract:
The first enantiocontrolled total synthesis of the marine sponge metabolite chlorodysinosin A is described. The structure and absolute configuration are identical to those of dysinosin A except for the presence of a novel 2S,3R-3-chloroleucine residue in the former. A concise stereocontrolled synthesis of the new chlorine-containing amino acid fragment was developed. An X-ray cocrystal structure of synthetic chlorodysinosin A with the enzyme thrombin confirms the structure and configuration assignment achieved through total synthesis. Within the aeruginosin family of natural products, chlorodysinosin A is the most potent inhibitor of the serine proteases thrombin, factor VIIa, and factor Xa, which are critical enzymes in the process leading to platelet aggregation and fibrin mesh formation in humans.
- Department of Chemistry, Université de Montréal, C.P. 6128, Station Centre-ville, Montréal, P.Q., H3C 3J7 Canada.
Organizational Affiliation: