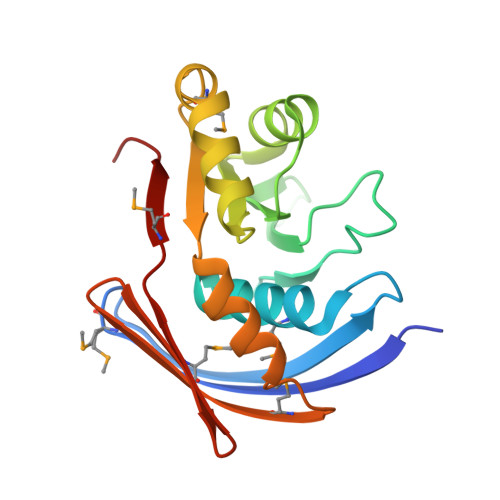
Structure of Xanthomonas axonopodis pv. citri YaeQ reveals a new compact protein fold built around a variation of the PD-(D/E)XK nuclease motif
Guzzo, C.R., Nagem, R.A.P., Barbosa, J.A.R.G., Farah, C.S.(2007) Proteins 69: 644-651
- PubMed: 17623842
- DOI: https://doi.org/10.1002/prot.21556
- Primary Citation of Related Structures:
2G3W - PubMed Abstract:
The YaeQ family of proteins are found in many Gram-negative and a few Gram-positive bacteria. We have determined the first structure of a member of the YaeQ family by X-ray crystallography. Comparisons with other structures indicate that YaeQ represents a new compact protein fold built around a variation of the PD-(D/E)XK nuclease motif found in type II endonucleases and enzymes involved in DNA replication, repair, and recombination. We show that catalytically important residues in the PD-(D/E)XK nuclease superfamily are spatially conserved in YaeQ and other highly conserved YaeQ residues may be poised to interact with nucleic acid structures.
- Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, Avenida Prof. Lineu Prestes 748, São Paulo, SP, CEP 05508-000, Brazil.
Organizational Affiliation: