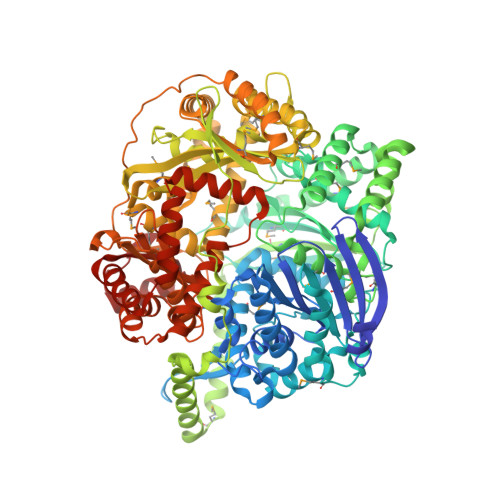
The closed structure of presequence protease PreP forms a unique 10 000 A(3) chamber for proteolysis
Johnson, K.A., Bhushan, S., Hallberg, B.M., Frohn, A., Glaser, E., Eneqvist, T.(2006) EMBO J 25: 1977-1986
- PubMed: 16601675
- DOI: https://doi.org/10.1038/sj.emboj.7601080
- Primary Citation of Related Structures:
2FGE - PubMed Abstract:
Presequence protease PreP is a novel protease that degrades targeting peptides as well as other unstructured peptides in both mitochondria and chloroplasts. The first structure of PreP from Arabidopsis thaliana refined at 2.1 Angstroms resolution shows how the 995-residue polypeptide forms a unique proteolytic chamber of more than 10,000 Angstroms(3) in which the active site resides. Although there is no visible opening to the chamber, a peptide is bound to the active site. The closed conformation places previously unidentified residues from the C-terminal domain at the active site, separated by almost 800 residues in sequence to active site residues located in the N-terminal domain. Based on the structure, a novel mechanism for proteolysis is proposed involving hinge-bending motions that cause the protease to open and close in response to substrate binding. In support of this model, cysteine double mutants designed to keep the chamber covalently locked show no activity under oxidizing conditions. The manner in which substrates are processed inside the chamber is reminiscent of the proteasome; therefore, we refer to this protein as a peptidasome.
- Department of Biochemistry and Biophysics, Arrhenius Laboratories for Natural Sciences, Stockholm University, Sweden.
Organizational Affiliation: