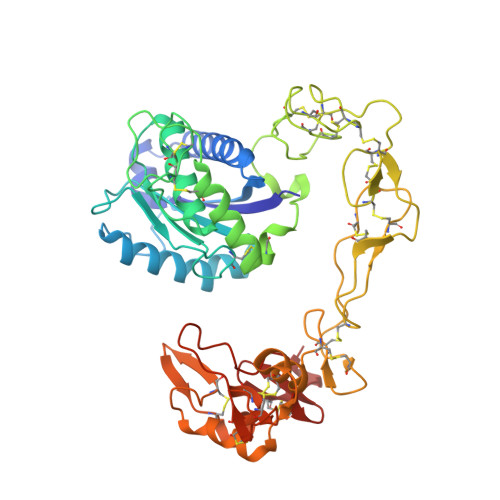
Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Igarashi, T., Araki, S., Mori, H., Takeda, S.(2007) FEBS Lett 581: 2416-2422
- PubMed: 17485084
- DOI: https://doi.org/10.1016/j.febslet.2007.04.057
- Primary Citation of Related Structures:
2DW0, 2DW1, 2DW2 - PubMed Abstract:
Catrocollastatin/vascular apoptosis-inducing protein (VAP)2B is a metalloproteinase from Crotalus atrox venom, possessing metalloproteinase/disintegrin/cysteine-rich (MDC) domains that bear the typical domain architecture of a disintegrin and metalloproteinase (ADAM)/adamalysin/reprolysin family proteins. Here we describe crystal structures of catrocollastatin/VAP2B in three different crystal forms, representing the first reported crystal structures of a member of the monomeric class of this family of proteins. The overall structures show good agreement with both monomers of atypical homodimeric VAP1. Comparison of the six catrocollastatin/VAP2B monomer structures and the structures of VAP1 reveals a dynamic, modular architecture that may be important for the functions of ADAM/adamalysin/reprolysin family proteins.
- Department of Cardiac Physiology, National Cardiovascular Center Research Institute 5-7-1 Fujishiro-dai, Suita, Osaka 565-8565, Japan.
Organizational Affiliation: