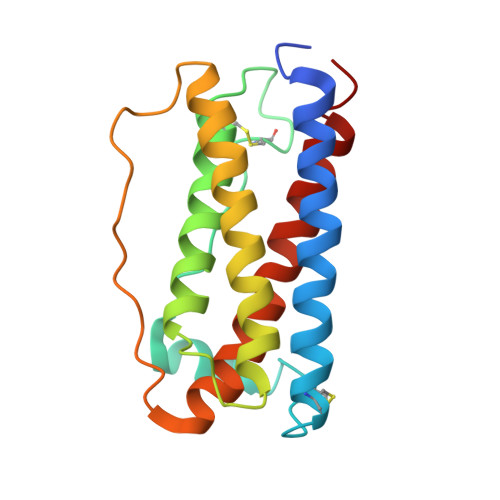
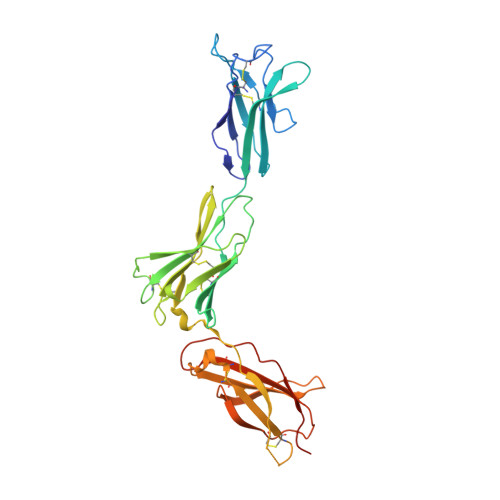
Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Tamada, T., Honjo, E., Maeda, Y., Okamoto, T., Ishibashi, M., Tokunaga, M., Kuroki, R.(2006) Proc Natl Acad Sci U S A 103: 3135-3140
- PubMed: 16492764
- DOI: https://doi.org/10.1073/pnas.0511264103
- Primary Citation of Related Structures:
2D9Q - PubMed Abstract:
A crystal structure of the signaling complex between human granulocyte colony-stimulating factor (GCSF) and a ligand binding region of GCSF receptor (GCSF-R), has been determined to 2.8 A resolution. The GCSF:GCSF-R complex formed a 2:2 stoichiometry by means of a cross-over interaction between the Ig-like domains of GCSF-R and GCSF. The conformation of the complex is quite different from that between human GCSF and the cytokine receptor homologous domain of mouse GCSF-R, but similar to that of the IL-6/gp130 signaling complex. The Ig-like domain cross-over structure necessary for GCSF-R activation is consistent with previously reported thermodynamic and mutational analyses.
- Research Group for Molecular Structural Biology, Quantum Beam Science Directorate, Japan Atomic Energy Agency, Tokai, Ibaraki.
Organizational Affiliation: