SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
Hayashi, T., Tanaka, Y., Sakai, N., Okada, U., Yao, M., Watanabe, N., Tamura, T., Tanaka, I.(2013) J Mol Biology 425: 3289-3300
- PubMed: 23831227
- DOI: https://doi.org/10.1016/j.jmb.2013.06.013
- Primary Citation of Related Structures:
2D6Y - PubMed Abstract:
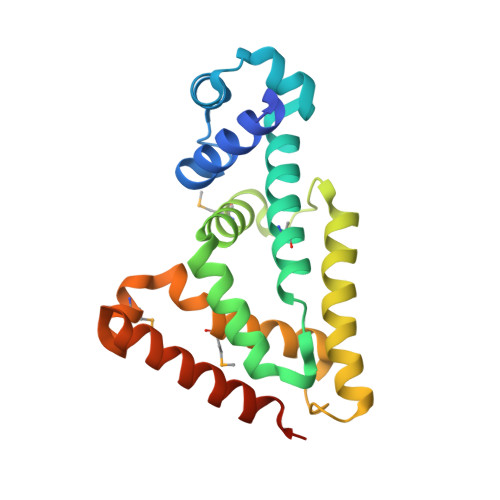
SCO4008 from Streptomyces coelicolor A3(2) is a member of the TetR family. However, its precise function is not yet clear. In this study, the crystal structure of SCO4008 was determined at a resolution of 2.3Å, and its DNA-binding properties were analyzed. Crystal structure analysis showed that SCO4008 forms an Ω-shaped homodimer in which the monomer is composed of an N-terminal DNA-binding domain containing a helix-turn-helix and a C-terminal dimerization and regulatory domain possessing a ligand-binding cavity. The genomic systematic evolution of ligands by exponential enrichment and electrophoretic mobility shift assay revealed that four SCO4008 dimers bind to the two operator regions located between sco4008 and sco4007, a secondary transporter belonging to the major facilitator superfamily. Ligand screening analysis showed that SCO4008 recognizes a wide range of structurally dissimilar cationic and hydrophobic compounds. These results suggested that SCO4008 is a transcriptional repressor of sco4007 responsible for the multidrug resistance system in S. coelicolor A3(2).
- Department of Food and Fermentation Science, Faculty of Food and Nutrition, Beppu University, Beppu, Oita 874-8501, Japan; Food Science and Nutrition, Graduate School of Food Science and Nutrition, Beppu University, Beppu, Oita 874-8501, Japan.
Organizational Affiliation: