Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
Papavoine, C.H., Christiaans, B.E., Folmer, R.H., Konings, R.N., Hilbers, C.W.(1998) J Mol Biology 282: 401-419
- PubMed: 9735296
- DOI: https://doi.org/10.1006/jmbi.1998.1860
- Primary Citation of Related Structures:
2CPB, 2CPS - PubMed Abstract:
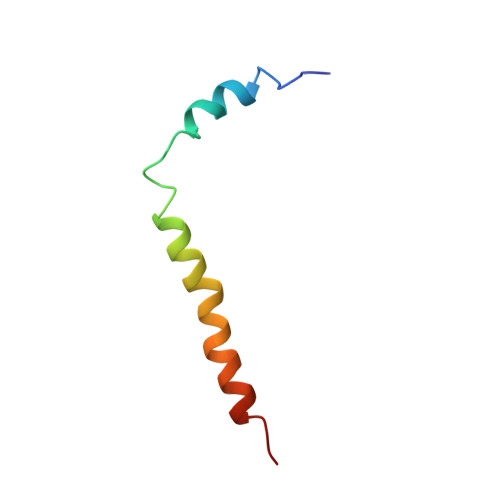
The three-dimensional structure of the major coat protein of bacteriophage M13, solubilized in detergent micelles, has been determined using heteronuclear multidimensional NMR and restrained molecular dynamics. The protein consists of two alpha-helices, running from residues 8 to 16 and 25 to 45, respectively. These two helices are connected by a flexible and distorted helical hinge region. The structural properties of the coat protein make it resemble a flail, in which the hydrophobic helix (residues 25 to 45) is the handle and the other, amphipathic, helix the swingle. In this metaphor, the hinge region is the connecting piece of leather. The mobility of the residues in the hinge region is likely to enable a smooth transformation from the membrane-bound form, mimicked by the structure in detergent micelles, into the structure in the mature phage. A specific distribution of the residues over the surface of the two helices was observed in the presented high-resolution structure of the membrane-bound form of the major coat protein as well as in the structure in the mature phage. All data suggest that this arrangement of residues is important for the interactions of the protein with the membrane, for correct protein-DNA and protein-protein interactions in the phage and for a proper growth of the phage during the assembly process. By combining our findings with earlier NMR results on the major coat protein in detergent micelles, we were able to construct a model that addresses the role of specific residues in the assembly process.
- Laboratory of Biophysical Chemistry, University of Nijmegen, Toernooiveld 6525 ED Nijmegen, The Netherlands.
Organizational Affiliation: