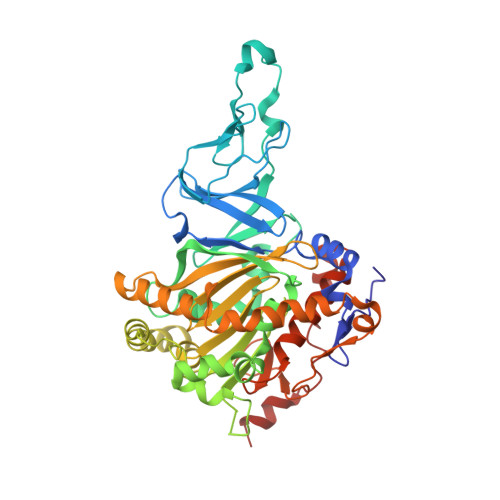
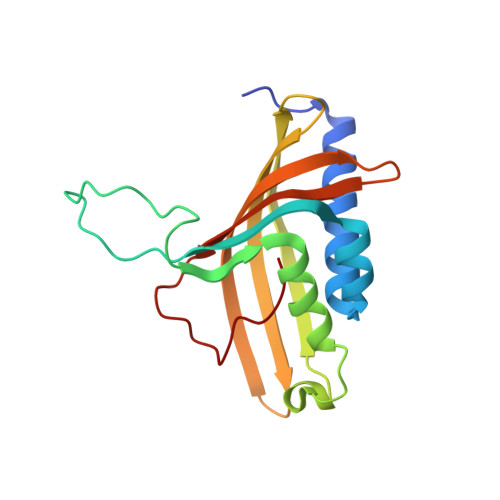
The Catalytic Pocket of the Ring-Hydroxylating Dioxygenase from Sphingomonas Chy-1.
Jakoncic, J., Jouanneau, Y., Meyer, C., Stojanoff, V.(2007) Biochem Biophys Res Commun 352: 861
- PubMed: 17157819
- DOI: https://doi.org/10.1016/j.bbrc.2006.11.117
- Primary Citation of Related Structures:
2CKF - PubMed Abstract:
Ring-hydroxylating dioxygenases are multicomponent bacterial enzymes that catalyze the first step in the oxidative degradation of aromatic hydrocarbons. The dioxygenase from Sphingomonas CHY-1 is unique in that it can oxidize a wide range of polycyclic aromatic hydrocarbons (PAHs). With a crystal structure similar to that of the seven other known dioxygenases, its catalytic domain features the largest hydrophobic substrate binding cavity characterized so far. Molecular modeling studies indicated that the catalytic cavity is large enough to accommodate a five-ring benzo[a]pyrene molecule. The predicted positions of this and other PAHs in the substrate binding pocket are consistent with the product regio- and stereo-selectivity of the enzyme.
- Brookhaven National Laboratory, National Synchrotron Light Source, Upton, NY 11973, USA.
Organizational Affiliation: