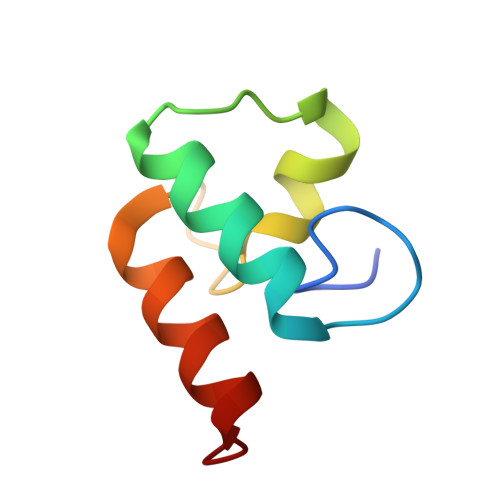
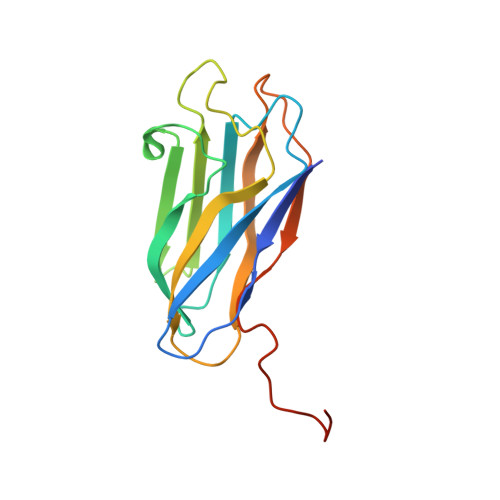Evidence for a Dual Binding Mode of Dockerin Modules to Cohesins.
Carvalho, A.L., Dias, F.M.V., Nagy, T., Prates, J.A.M., Proctor, M.R., Smith, N., Bayer, E.A., Davies, G.J., Ferreira, L.M.A., Romao, M.J., Fontes, C.M.G.A., Gilbert, H.J.(2007) Proc Natl Acad Sci U S A 104: 3089
- PubMed: 17360613
- DOI: https://doi.org/10.1073/pnas.0611173104
- Primary Citation of Related Structures:
2CCL - PubMed Abstract:
The assembly of proteins that display complementary activities into macromolecular complexes is critical to cellular function. One such enzyme complex, of environmental significance, is the plant cell wall degrading apparatus of anaerobic bacteria, termed the cellulosome. The complex assembles through the interaction of enzyme-derived "type I dockerin" modules with the multiple "cohesin" modules of the scaffolding protein. Clostridium thermocellum type I dockerin modules contain a duplicated 22-residue sequence that comprises helix-1 and helix-3, respectively. The crystal structure of a C. thermocellum type I cohesin-dockerin complex showed that cohesin recognition was predominantly through helix-3 of the dockerin. The sequence duplication is reflected in near-perfect 2-fold structural symmetry, suggesting that both repeats could interact with cohesins by a common mechanism in wild-type (WT) proteins. Here, a helix-3 disrupted mutant dockerin is used to visualize the reverse binding in which the dockerin mutant is indeed rotated 180 degrees relative to the WT dockerin such that helix-1 now dominates recognition of its protein partner. The dual binding mode is predicted to impart significant plasticity into the orientation of the catalytic subunits within this supramolecular assembly, which reflects the challenges presented by the degradation of a heterogeneous, recalcitrant, insoluble substrate by a tethered macromolecular complex.
- Rede de Química e Tecnologia/Centro de Química Fina e Biotecnologia (REQUIMTE/CQFB), Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal.
Organizational Affiliation: