Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
Ekici, O.D., Li, Z.Z., Campbell, A.J., James, K.E., Asgian, J.L., Mikolajczyk, J., Salvesen, G.S., Ganesan, R., Jelakovic, S., Grutter, M.G., Powers, J.C.(2006) J Med Chem 49: 5728
- PubMed: 16970398
- DOI: https://doi.org/10.1021/jm0601405
- Primary Citation of Related Structures:
2C1E, 2C2K, 2C2M, 2C2O, 2C2Z - PubMed Abstract:
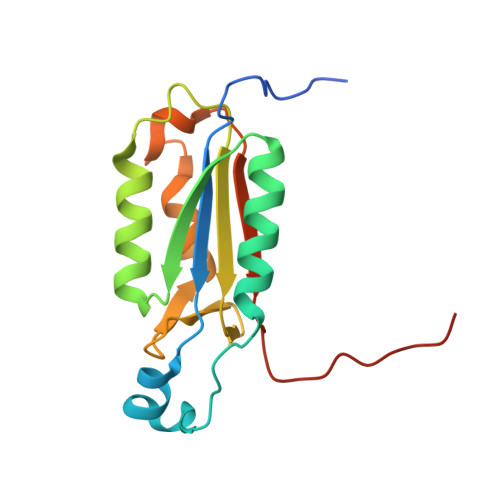
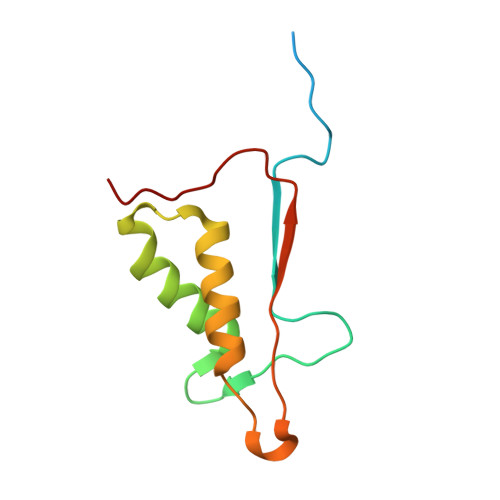
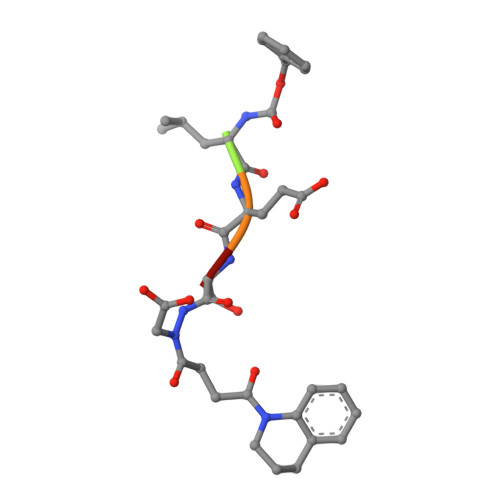
Aza-peptide Michael acceptors are a novel class of inhibitors that are potent and specific for caspases-2, -3, -6, -7, -8, -9, and -10. The second-order rate constants are in the order of 10(6) M(-1) s(-1). The aza-peptide Michael acceptor inhibitor 18t (Cbz-Asp-Glu-Val-AAsp-trans-CH=CH-CON(CH(2)-1-Naphth)(2) is the most potent compound and it inhibits caspase-3 with a k(2) value of 5620000 M(-1) s(-1). The inhibitor 18t is 13700, 190, 6.4, 594, 37500, and 173-fold more selective for caspase-3 over caspases-2, -6, -7, -8, -9, and -10, respectively. Aza-peptide Michael acceptors designed with caspase specific sequences are selective and do not show any cross reactivity with clan CA cysteine proteases such as papain, cathepsin B, and calpains. High-resolution crystal structures of caspase-3 and caspase-8 in complex with aza-peptide Michael acceptor inhibitors demonstrate the nucleophilic attack on C2 and provide insight into the selectivity and potency of the inhibitors with respect to the P1' moiety.
- School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia 30332-0400, USA.
Organizational Affiliation: