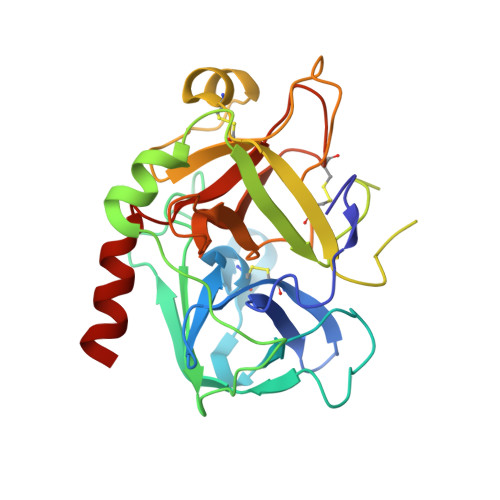
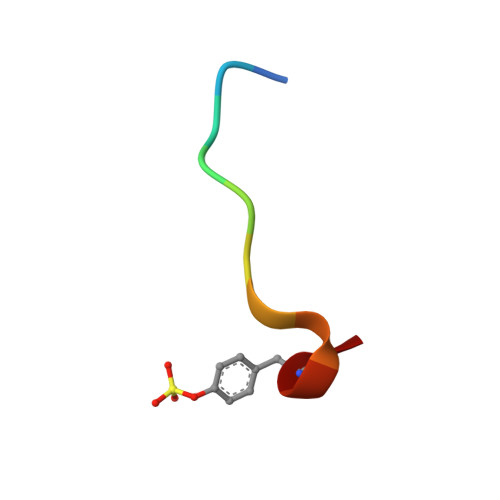
Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Bulat, S., Bosio, S., Papadopoulos, M.A., Cerezo-Galvez, S., Grabowski, E., Rosenbaum, C., Matassa, V.G., Ott, I., Metz, G., Schamberger, J., Sekul, R., Feurer, A.(2006) Lett Drug Des Discov 3: 289