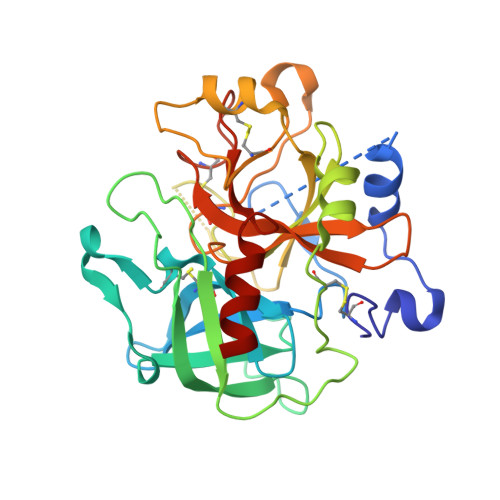
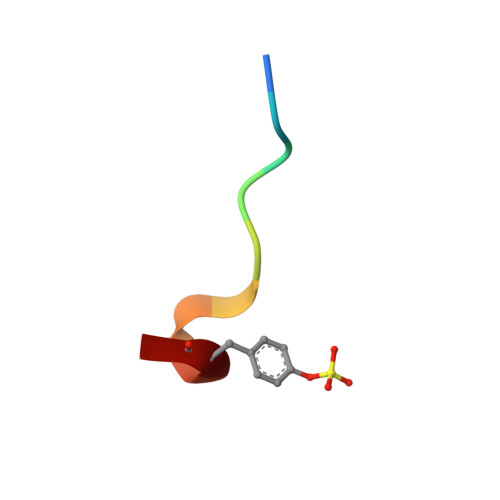
Phenolic P2/P3 core motif as thrombin inhibitors--design, synthesis, and X-ray co-crystal structure.
Hanessian, S., Therrien, E., van Otterlo, W.A., Bayrakdarian, M., Nilsson, I., Xue, Y.(2006) Bioorg Med Chem Lett 16: 1032-1036
- PubMed: 16290930
- DOI: https://doi.org/10.1016/j.bmcl.2005.10.082
- Primary Citation of Related Structures:
2BDY - PubMed Abstract:
Prototypical thrombin inhibitors were synthesized based on a trisubstituted phenol as a core motif. A naphthylsulfonamide analogue showed excellent antithrombin activity. An X-ray co-crystal structure showed the expected interactions.
- Department of Chemistry, Université de Montréal, C.P. 6128, Succ. Centre-Ville, Montréal, PQ, Canada H3C 3J7. stephen.hanessian@umontreal.ca
Organizational Affiliation: