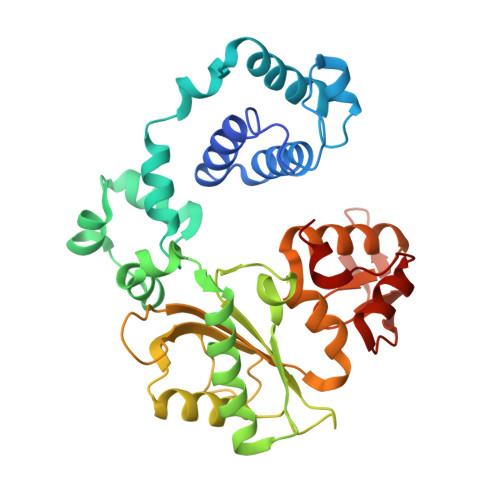
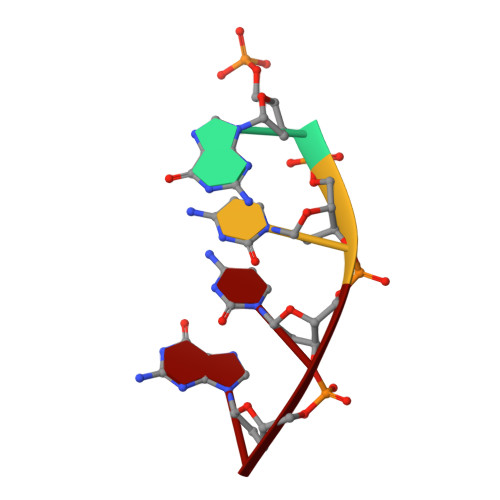
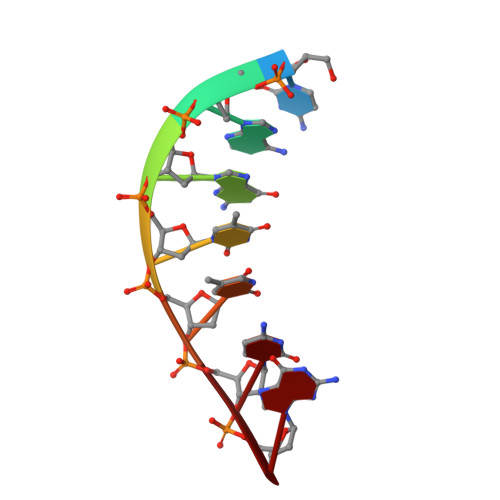
Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Garcia-Diaz, M., Bebenek, K., Krahn, J.M., Pedersen, L.C., Kunkel, T.A.(2006) Cell 124: 331-342
- PubMed: 16439207
- DOI: https://doi.org/10.1016/j.cell.2005.10.039
- Primary Citation of Related Structures:
2BCQ, 2BCR, 2BCS, 2BCU, 2BCV - PubMed Abstract:
Insertions and deletions in coding sequences can alter the reading frame of genes and have profound biological consequences. In 1966, Streisinger proposed that these mutations result from strand slippage, which in repetitive sequences generates misaligned intermediates stabilized by correct base pairing that support polymerization. We report here crystal structures of human DNA polymerase lambda, which frequently generates deletion mutations, bound to such intermediates. Each contains an extrahelical template nucleotide upstream of the active site. Surprisingly, the extra nucleotide, even when combined with an adjacent mismatch, does not perturb polymerase active site geometry, which is indistinguishable from that for correctly aligned strands. These structures reveal how pol lambda can polymerize on substrates with minimal homology during repair of double-strand breaks and represent strand-slippage intermediates consistent with Streisinger's classical hypothesis. They are thus relevant to the origin of single-base deletions, a class of mutations that can confer strong biological phenotypes.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: