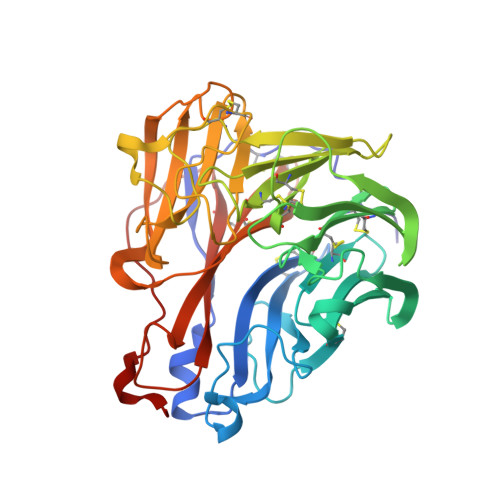
The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor.
Varghese, J.N., McKimm-Breschkin, J.L., Caldwell, J.B., Kortt, A.A., Colman, P.M.(1992) Proteins 14: 327-332
- PubMed: 1438172
- DOI: https://doi.org/10.1002/prot.340140302
- Primary Citation of Related Structures:
2BAT - PubMed Abstract:
Crystallographic studies of neuraminidase-sialic acid complexes indicate that sialic acid is distorted on binding the enzyme. Three arginine residues on the enzyme interact with the carboxylate group of the sugar which is observed to be equatorial to the saccharide ring as a consequence of its distorted geometry. The glycosidic oxygen is positioned within hydrogen-bonding distance of Asp-151, implicating this residue in catalysis.
- CSIRO, Division of Biomolecular Engineering, Parkville, Victoria, Australia.
Organizational Affiliation: