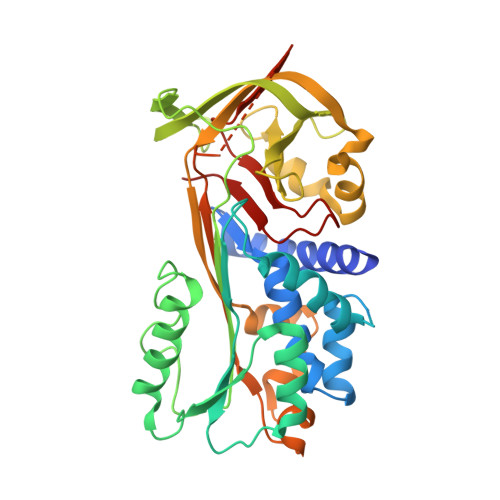
Crystal structure of SCCA1 and insight about the interaction with JNK1
Zheng, B., Matoba, Y., Kumagai, T., Katagiri, C., Hibino, T., Sugiyama, M.(2009) Biochem Biophys Res Commun 380: 143-147
- PubMed: 19166818
- DOI: https://doi.org/10.1016/j.bbrc.2009.01.057
- Primary Citation of Related Structures:
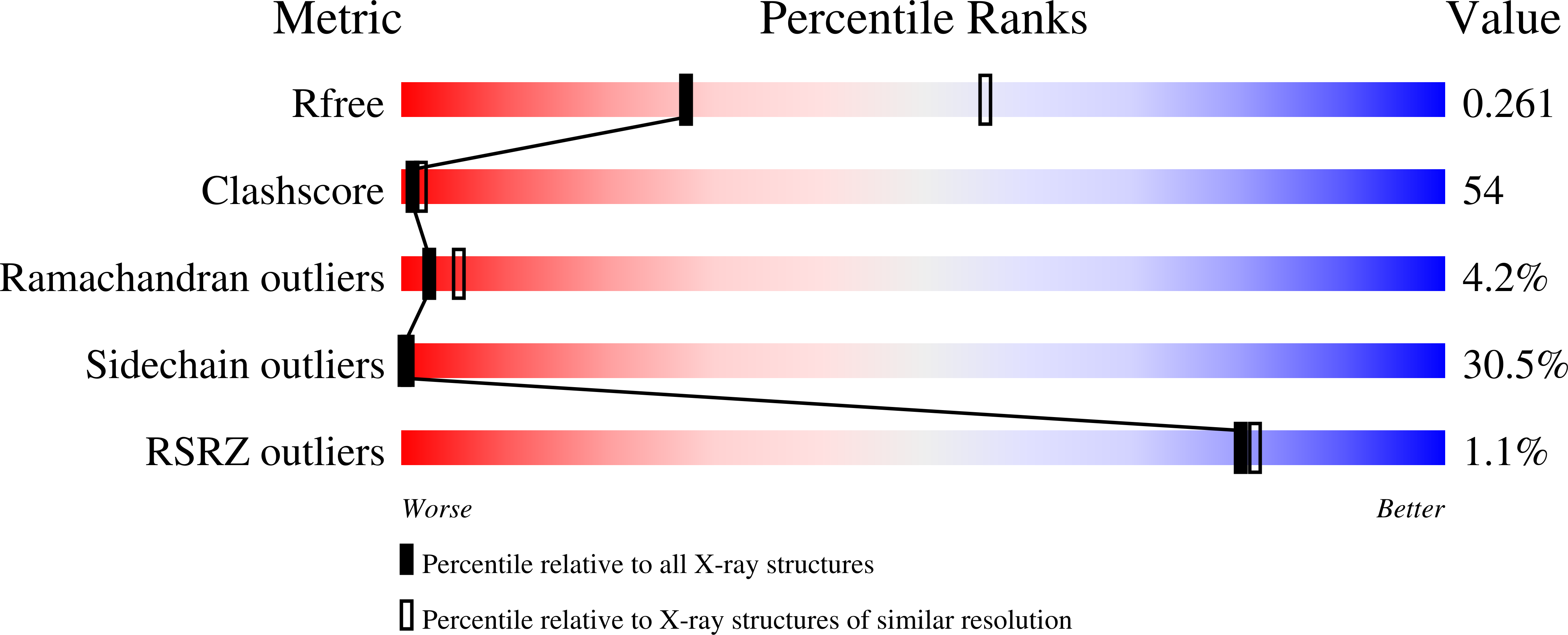
2ZV6 - PubMed Abstract:
Squamous cell carcinoma antigen 1 (SCCA1), which belongs to serine proteinase inhibitor (serpin) superfamily, inhibits papain-like cysteine proteinase. Recently, it has been reported that SCCA1 acts not only as a proteinase inhibitor but also as an inhibitor of UV-induced apoptosis via suppression of the activity of c-Jun NH(2)-terminal kinase (JNK1). The present study determined the crystal structure of SCCA1, suggesting that the reactive center loop (RCL) of SCCA1, a recognition site of proteinase, is very flexible and located away form the main-body of SCCA1. We show that the inhibitory effect of SCCA1 on the kinase activity of JNK1 is lost when the RCL was truncated. Furthermore, we found that a mutant protein created by replacing one amino acid in RCL maintain the suppressive activity to JNK1, whereas the inhibitory effect to proteinase is obviously decreased.
- Department of Molecular Microbiology and Biotechnology, Graduate School of Biomedical Sciences, Hiroshima University, Kasumi 1-2-3, Minami-Ku, Hiroshima 734-8551, Japan.
Organizational Affiliation: