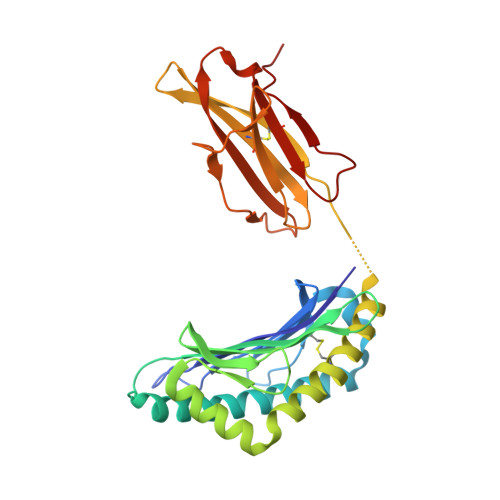
Prevention of cytotoxic T cell escape using a heteroclitic subdominant viral T cell determinant.
Butler, N.S., Theodossis, A., Webb, A.I., Nastovska, R., Ramarathinam, S.H., Dunstone, M.A., Rossjohn, J., Purcell, A.W., Perlman, S.(2008) PLoS Pathog 4
- PubMed: 18949029
- DOI: https://doi.org/10.1371/journal.ppat.1000186
- Primary Citation of Related Structures:
2ZSV, 2ZSW - PubMed Abstract:
High affinity antigen-specific T cells play a critical role during protective immune responses. Epitope enhancement can elicit more potent T cell responses and can subsequently lead to a stronger memory pool; however, the molecular basis of such enhancement is unclear. We used the consensus peptide-binding motif for the Major Histocompatibility Complex molecule H-2K(b) to design a heteroclitic version of the mouse hepatitis virus-specific subdominant S598 determinant. We demonstrate that a single amino acid substitution at a secondary anchor residue (Q to Y at position 3) increased the stability of the engineered determinant in complex with H-2K(b). The structural basis for this enhanced stability was associated with local alterations in the pMHC conformation as a result of the Q to Y substitution. Recombinant viruses encoding this engineered determinant primed CTL responses that also reacted to the wildtype epitope with significantly higher functional avidity, and protected against selection of virus mutated at a second CTL determinant and consequent disease progression in persistently infected mice. Collectively, our findings provide a basis for the enhanced immunogenicity of an engineered determinant that will serve as a template for guiding the development of heteroclitic T cell determinants with applications in prevention of CTL escape in chronic viral infections as well as in tumor immunity.
- Department of Microbiology, University of Iowa, Iowa City, Iowa, United States of America.
Organizational Affiliation: