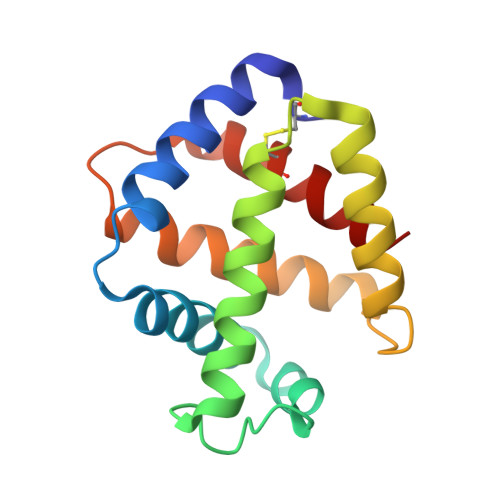
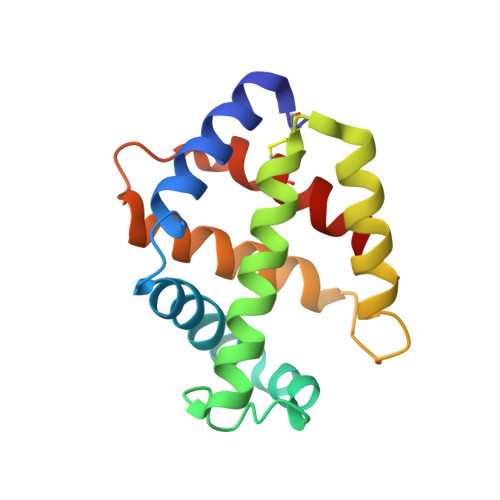
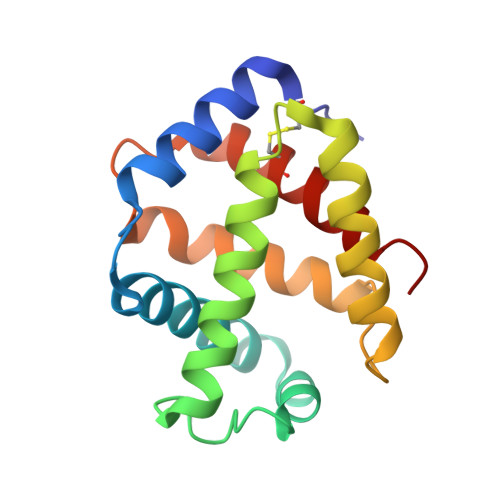
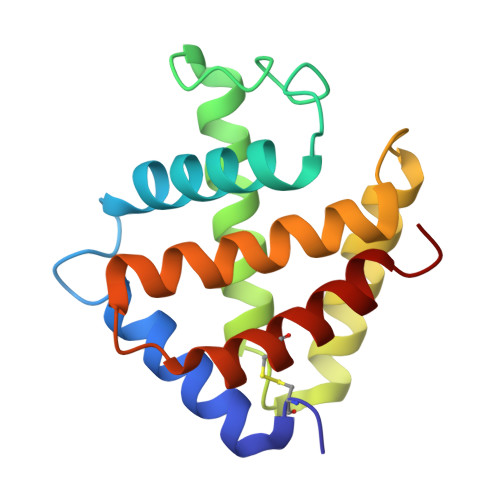
Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin
Numoto, N., Nakagawa, T., Kita, A., Sasayama, Y., Fukumori, Y., Miki, K.(2008) Biochemistry 47: 11231-11238
- PubMed: 18834142
- DOI: https://doi.org/10.1021/bi8012609
- Primary Citation of Related Structures:
2ZS0, 2ZS1 - PubMed Abstract:
The oxygen binding properties of extracellular giant hemoglobins (Hbs) in some annelids exhibit features significantly different from those of vertebrate tetrameric Hbs. Annelid giant Hbs show cooperative oxygen binding properties in the presence of inorganic cations, while the cooperativities of vertebrate Hbs are enhanced by small organic anions or chloride ions. To elucidate the structural basis for the cation-mediated cooperative mechanisms of these giant Hbs, we determined the crystal structures of Ca2+- and Mg2+-bound Hbs from Oligobrachia mashikoi at 1.6 and 1.7 A resolution, respectively. Both of the metal-bound structures were determined in the oxygenated state. Four Ca2+-binding sites and one Mg2+-binding site were identified in each tetramer subassembly. These cations are considered to stabilize the oxygenated form and increase affinity and cooperativity for oxygen binding, as almost all of the Ca2+ and Mg2+ cations were bound at the interface regions, forming either direct or hydrogen bond-mediated interactions with the neighboring subunits. A comparison of the structures of the oxygenated form and the partially unliganded form provides structural insight into proton-coupled cooperativity (Bohr effect) and ligand-induced transitions. Two histidine residues are assumed to be primarily associated with the Bohr effect. With regard to the ligand-induced cooperativity, a novel quaternary rotation mechanism is proposed to exist at the interface region of the dimer subassembly. Interactions among conserved residues Arg E10, His F3, Gln F7, and Val E11, together with the bending motion of the heme molecules, appear to be essential for quaternary rearrangement.
- Department of Chemistry, Graduate School of Science, Kyoto UniVersity, Sakyo-ku, Kyoto 606-8502, Japan.
Organizational Affiliation: